Arrayed CRISPR Libraries
transEDIT Arrayed CRISPR Libraries
- Enabling single-target or combinatorial vector-based genetic screens at scale in arrayed format
- sgRNAs designed by CRoatan algorithm – read the publication in Molecular Cell
- Highly efficient lentiviral vector backbone for producing high-titer lentiviral particles
- Exceptionally bright ZsGreen fluorescent reporter
- All CRISPR library sgRNA constructs are 100% sequence-confirmed
- Order premade or custom arrayed CRISPR Libraries
transEDIT CRISPR Arrayed Libraries allow single-target or combinatorial genetic screens to be carried out at scale in a multiplexed or arrayed format. sgRNAs are designed using the CRoatan algorithm which is based on a machine-learning approach combined with other strategies to optimize knockout efficiency with the CRISPR/Cas9 system.
The only commercially available Dual sgRNA Arrayed CRISPR KO Library:
The transEDIT Dual sgRNA Arrayed CRISPR KO Library covering the Human Genome was developed by transOMIC in partnership with Cold Spring Harbor Laboratory using the CRoatan algorithm and a multiplexed sgRNA expression for combinatorial targeting. These strategies have been combined to design and construct a human genome-wide, sequence-verified, arrayed CRISPR library, employing the enhancement effect of co-expressing two different sgRNAs for the same target:
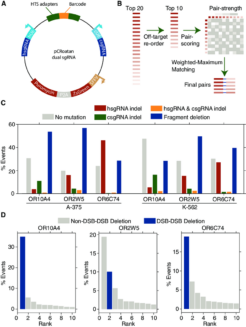
Dual sgRNA Arrayed CRISPR KO Library: Targeting with Multiple sgRNAs Results in Predictable Genomic Scars
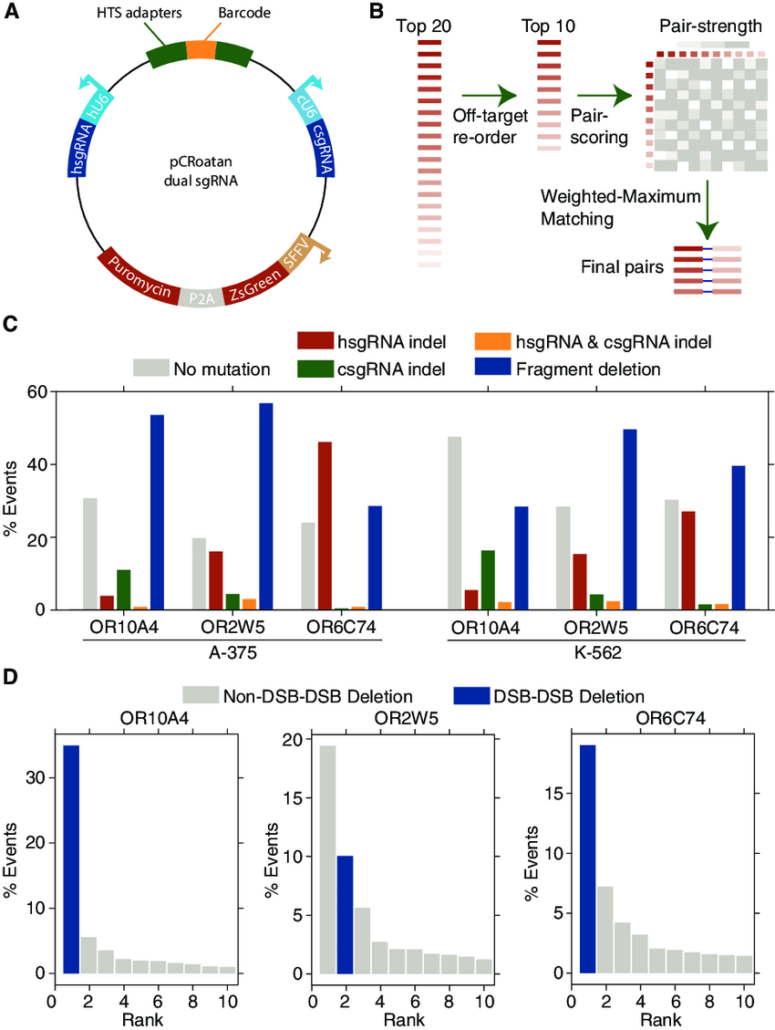
(A) Schematic map of the lentiviral, dual-sgRNA expression vector with relevant features highlighted. hU6, human U6 promoter; cU6, chicken U6 promoter; hsgRNA, human U6-promoterdriven sgRNA; csgRNA, human U6-promoterdriven sgRNA; HTS, high-throughput sequencing adapters; SFFV, spleen focus-forming virus promoter. (B) sgRNA pairing algorithm used to design five targeting constructs for a gene. The top 20 sgRNAs for each gene are filtered to a set of 10 to reduce the probability of off-targeting effects. Pairs within these 10 are then scored using the set of heuristics defined in Figure S2. The resultant pairing matrix is then used as input for a maximum-weighted matching algorithm to define a final set of 5 sgRNA pairs. (C) Paired-end sequencing analysis of genomic scars left after dual-CRoatan NEG-targeting constructs have been infected into A-375 and K-562 cells. hsgRNA and csgRNA indels are where only one of the two targeted regions shows mutational burden in an HTS fragment. hsgRNA and csgRNA indel counts represent cases where both targets have indels, and fragment deletions are where the region between the two targets is deleted. (D) Analysis of the genomic scars described in (C) that correspond to fragment deletions between two sgRNA target sites. The top ten most frequent deletions are shown with their corresponding rate of occurrence, as measured by their average frequency in infected A-375 and K-562 cells. Scars that result from exact deletion of the doublestrand-break-flanked fragment are annotated as DSB-DSB deletions. Scars where, in addition to the fragment deletion, other bases are inserted or deleted are annotated as non-DSB-DSB deletions.
You can purchase the whole genome wide arrayed dual sgRNA library (Click here to download the gene list) or you can pick and choose for building a custom arrayed library.
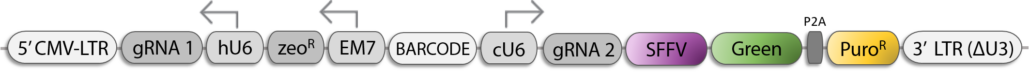
Dual sgRNA Lentiviral Vector for CRISPR KO
transEDIT Custom Arrayed CRISPR Libraries: Any size – Any application
Our partner transOMIC Technologies offers maximal flexibility for your arrayed CRISPR screening project, based on their wealth of vectorology, and clone handling experience.
transOMIC pCLIP CRISPR Vectors:
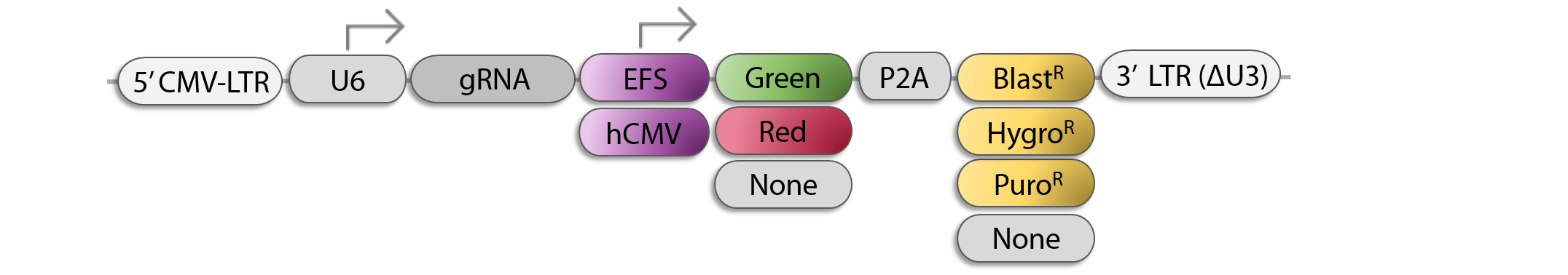
Single sgRNA Lentiviral Vector Options for CRISPR KO, CRISPRa, or CRISPRi
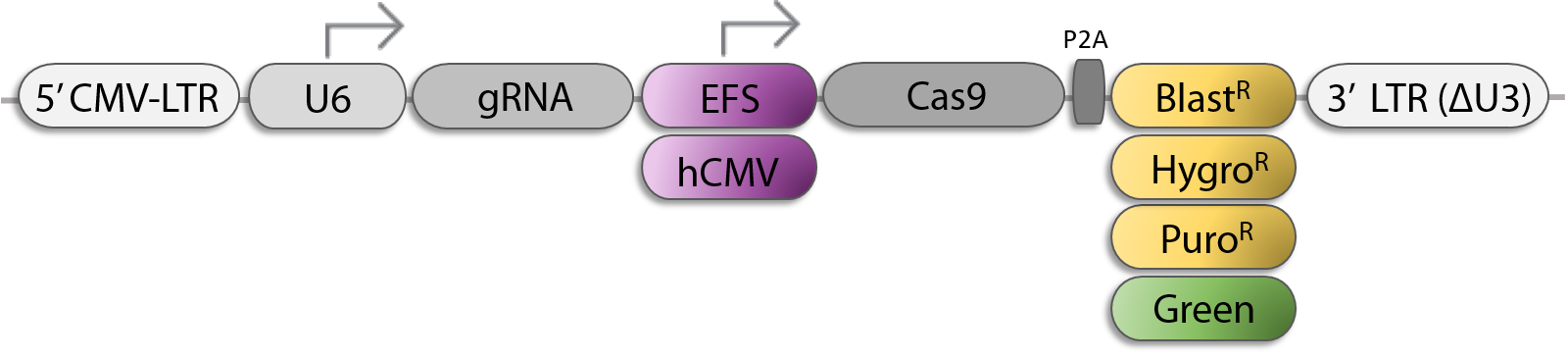
All-In-One sgRNA/Cas9 Lentiviral Vector Options for CRISPR KO
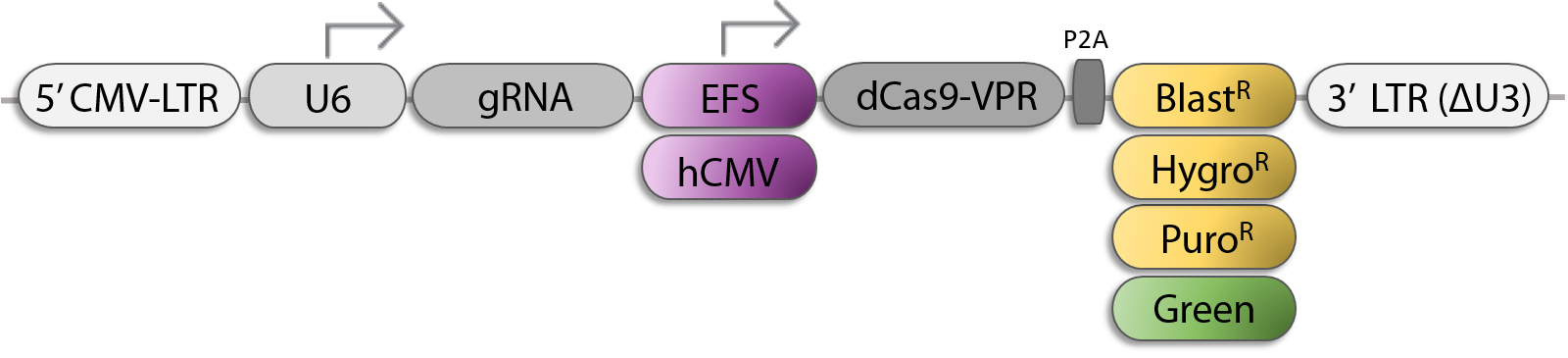
All-In-One sgRNA/Cas9-VPR Lentiviral Vector Options for CRISPRa
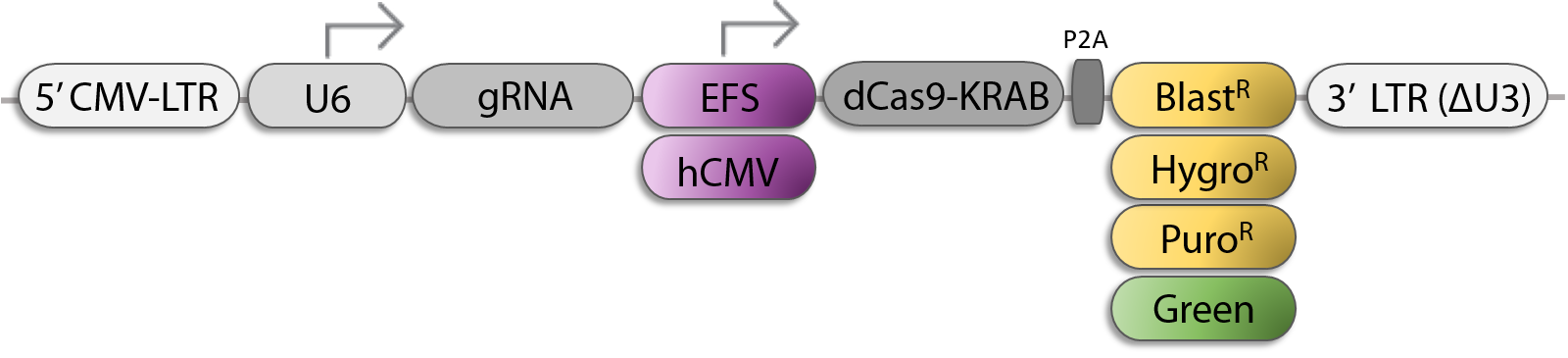
All-In-One sgRNA/Cas9-KRAB Lentiviral Vector Options for CRISPRi
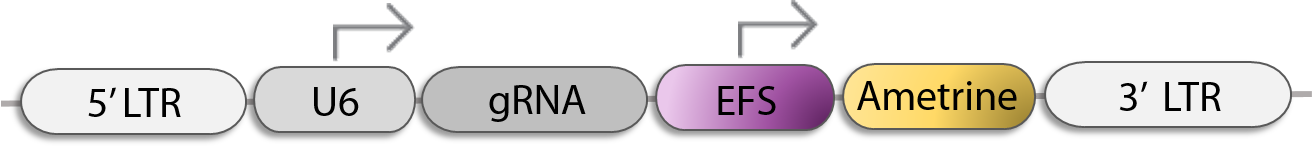
Single sgRNA Retroviral Vector for CRISPR KO, CRISPRa, or CRISPRi
- Build your custom CRISPR array cloned in any of transOMIC´s pCLIP lentiviral/retroviral vectors for CRISPR KO, CRISPRa, or CRISPRi. If you prefer your own lentiviral vector, we can accomodate this, too!
- sgRNAs for your target gene list will be designed using the CRoatan algorithm or you can share your own sgRNA designs
- Oligos are synthesized and cloned into your choice of lentiviral vector.
- Each clone is sequence-verified and arrayed into 96-well plates.
- We can deliver glycerol stocks, plasmid DNA, and/or ready-to-use lentiviral particles in 96-well plates.
Please contact us with your project details and share your gene list, the number of sgRNAs needed for targeting each gene, and your vector choice with us. We will be pleased to send you a detailed quotation.
Showing all 5 results