Description
SHAPE RNA Structure Probing for Single RNA
An easy and fast protocol that allows the user to obtain in vitro SHAPE RNA Structure Probing data by NGS through NAI structure probing of an IVT RNA
eSHAPE single RNA is a method to obtain SHAPE RNA structure probing data for in vitro transcribed RNA formulations. In vitro transcribed or synthetic RNAs are used as input for RNA structure probing with the NAI reagent. eSHAPE single RNA is available as a kit for end user completion, and as a full service offering with results delivered as a data package containing RNA structure information as reactivity values.
• Compare similar single RNAs to identify regions of RNA folding variability
• Reveal differences in RNA folding across different conditions or due to RNA binding factors
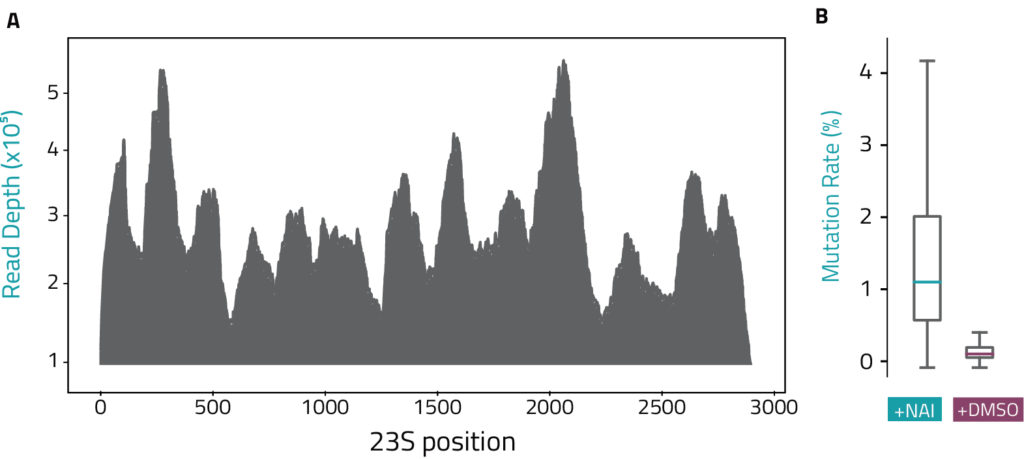
• Extremely deep RNA coverage >10,000X with minimal sequencing depth
• Elevated mutation rates in NAI samples compared to the DMSO control identify RNA positions with high reactivity/unpaired bases
• Reactivity values are predictive of paired and unpaired base status in an RNA secondary structure fold
Eclipse Bio´s SHAPE RNA Structure Probing technology on IVT RNAs measures local RNA flexibility in vitro for prediction of RNA secondary structure for the RNA formulation:
Selective 2’-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) is a chemical probing method that measures RNA flexibility at single nucleotide resolution. The licensed use of the patented reagent NAI (2-methylnicotinic acid imidazolide) in the RNA probing reaction forms adducts with the free 2′-OH on the RNA backbone in single stranded regions. During the reverse transcription reaction these adducts induce mutations in the newly formed cDNA at a higher rate than the DMSO control sample. Mutation rates are determined by NGS and calculated for each position along the IVT RNA. Computing into SHAPE reactivities guides the folding of the RNA by indicating the pairing status for each position.
Sample Input: 1 ug in vitro transcribed or synthetic RNA
Sequencing Recommendations: 8M reads (SE100) on Illumina
Kit contents:
– RNA probing reagents incl. NAI for 8 samples
– DMSO for 8 control samples
– All enzymes, primers, adapters, and buffers needed to process 8 samples/to construct 16 NGS Libraries (8 NAI libraries and 8 DMSO libraries)
– Size selection beads
– Index primers
The Eclipse BioInnovations eSHAPE Single RNA Data Analysis Package delivers the results using Fastq files of the raw sequencing reads for each NAI library and DMSO library obtained by you as follows:
FASTA
BAM
SHAPE
QC Metrics
Coverage Plots
Mutation Rate Plots
Reactivity Plots
Reactivity Comparison Plots
Statistical Report