Description
m6A-eCLIP: Enhanced meRIP-seq for superior m6A-seq
Efficient and precise identification of RNA m6A modifications by superior m6A-seq combining methylated RNA immunprecipitation sequencing meRIP-seq and the enhanced CLIP-seq protocol
Eclipse BioInnovation´s m6A-eCLIP library preparation kit for m6A-seq combines the use of an m6A antibody with the eCLIP technology, based on Van Nostrand, Yeo et al. (Nature Methods, 2016 June; 13(6): 508–514.) which results in an enhanced meRIP-seq protocol.
• More sites with less input: The m6A-eCLIP high throughput m6A-seq method enables profiling m6A marks with 100-fold less mRNA input than conventional approaches such as meRIP-seq
• Precise m6A site resolution: Single-nucleotide modification site calling
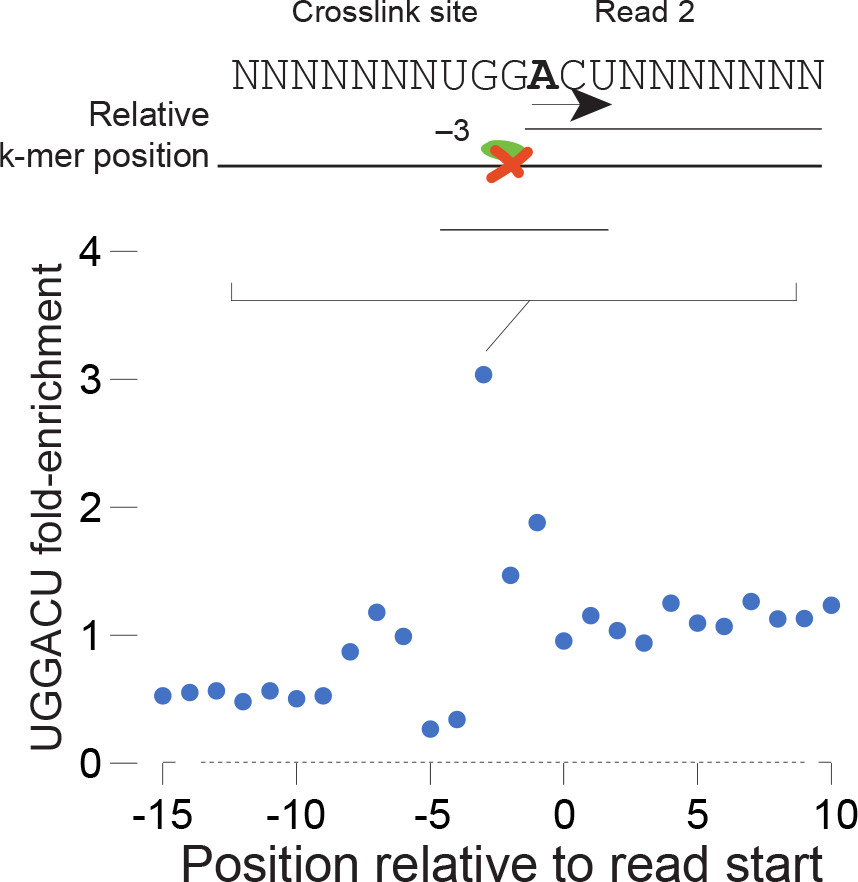
• Unbiased with high specificity: m6A eCLIP captures the DRACH motif as well as enriched modifications near stop codons and in the 3´UTRs
N6-methyladenosine (m6A) is the most prevalent epitranscriptomic RNA modification found in eukaryotes, and m6A has been shown to regulate many stages of RNA biology including splicing, secondary/tertiary structure, localization, stability, and translation. Adenosine modifications are actively added, recognized, and removed by a series of “writer”, “reader”, and “eraser” proteins, making m6A a dynamic and reversible system for regulating RNAs.
The m6A-eCLIP Kit is a targeted RNA modification sequencing kit that has applications in many fields of research. Starting with purified RNA, the kit allows researchers to globally map m6A RNA modification states, which are associated with cancer, autoimmune disease, obesity, and other disease indications.Acquisition of host-like m6A by viral RNA also plays a role in evading detection by the innate immune system.
The kit combines the use of an m6A antibody with the eCLIP technology (Van Nostrand et al., Nature Methods, 2016): RNA is chemically fragmented into 100 nucleotides or smaller fragments, followed by magnetic immunoprecipitation with an m6A antibody. After immunoprecipitation, isolated RNA fragments are subjected to library preparation including adapter ligation, reverse transcription, and PCR amplification. The streamlined 4-5 day protocol is easy to process and produces high quality libraries, and can be broken down into multiple start and stop steps. The kit enables the user to do mRNA isolation, m6A IP and library prep on up to 8 high quality samples (8 IPs and 8 inputs).
The Eclipse BioInnovations m6AeCLIP-Seq Data Analysis Package includes peak calling and input normalization using Fastq files of the raw sequencing reads obtained by you. An HTML report detailing the figures related to all significantly enriched A methylation peaks and containing the location of each input-normalized A-methylation peak in the genome will be generated.