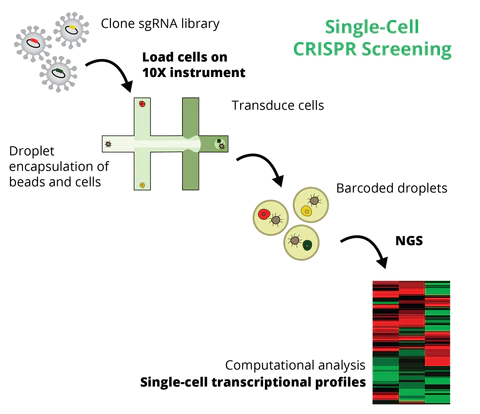
Pooled Perturb-Seq CRISPR KO Libraries
Combine pooled CRISPR genetic screens with RNA expression profiling at the single-cell level
About Perturb-Seq
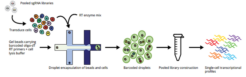
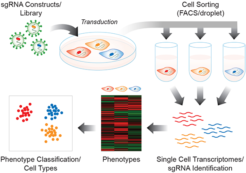
Perturb-Seq (aka CROP-Seq or CRISPR-seq) is a single-cell functional genomics method that combines pooled CRISPR (sgRNA) genetic screening with single-cell expression analysis. It is a promising technology for the efficient and scalable interrogation of many individual genes in one single experiment, enabling the identification of transcriptional profiles linked to specific gene-KOs.
Changes in gene expression associated with specific sgRNAs can be directly linked to specific gene knockouts.
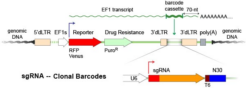
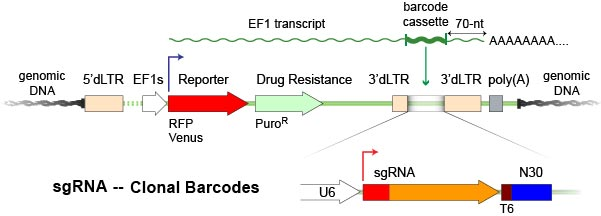
CROP-Seq CRISPR Libraries in pScribeA barcoded sgRNA expression cassette is inserted into the 3’-region of an expressed selection marker gene on Cellecta´s pScribe vector. The guide is expressed near the poly-A+ tail of a standard mRNA transcript so it is reverse-transcribed and detectable in the cDNA generated as part of the RNA-seq protocol. |
 |
Our partner Cellecta can provide CRISPR sgRNA libraries that accommodate any scRNA system.
Besides Crop-Seq libraries in pScribe, Nucleus Biotech provide any variation of sgRNA libraries or other customized designs for your list of target genes as part of Cellecta´s custom library design service.
You may also be interested in premade, ready-to-use CloneTracker XP CRISPR Knockout libraries, targeting human and mouse anti-cancer genes. These libraries express a sgRNA for pooled CRISPR genetic screens with a unique molecular identifier (UMI or barcode) that facilitates clonal cell tracking and single-cell expression analysis.
sgRNA and barcode can be detected in the RNA expression data.
Contact us for a quotation.
Showing all 10 results