RNA Binding Protein Target Sequencing: Enhanced CLIP-Seq
Revolutionize the ability to identify RBP binding sites
The processing of and regulation through RNA molecules is tightly controlled by RNA binding proteins (RBPs), which bind to RNAs in trans through recognition of sequence and structural motifs and regulate RNA processing in cell-type, condition-specific, or temporal manners.
Recent studies have estimated over 1500 RBPs in the human genome, which play roles throughout the RNA life cycle. Mutation of proper RBP activity has been linked to cancer, Amyotrophic Lateral Sclerosis (ALS), and numerous other diseases, and the emergence of new sequencing techniques will continue to rapidly expand our knowledge of RBPs causally mutated in diseases.
The eCLIP-seq method
Enhanced crosslinking by UV and immunoprecipitation followed by high-throughput sequencing (eCLIP-seq, van Nostrand et al. Nature Methods 2016 and Nature 2020) was developed to provide a robust and reproducible framework to identify RNA binding protein targets.
eCLIP is based on crosslinking RBP-RNA interactions by UV and the selective transfer of only the crosslinked RNA fragments to a nitrocellulose membrane. The succeeding library preparation method employs a highly efficient adapter ligation strategy as well as Unique Molecular Identifiers (UMIs) to ensure highest quantification accuracy.
- Increased efficiency, decreased PCR duplication
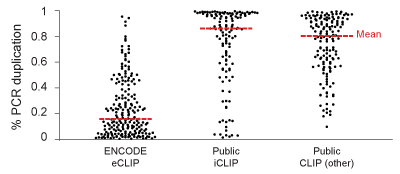
eCLIP-seq builds upon improved library preparation efficiency by nearly one thousand-fold based on much better adapter ligation chemistries. This increases experimental success rates and decreases wasted sequencing due to PCR duplication, see above: 102 eCLIP experiments show lower percentage of PCR duplications, relative to published iCLIP and other CLIP datasets
- Transcriptome-wide identification of RNA targets
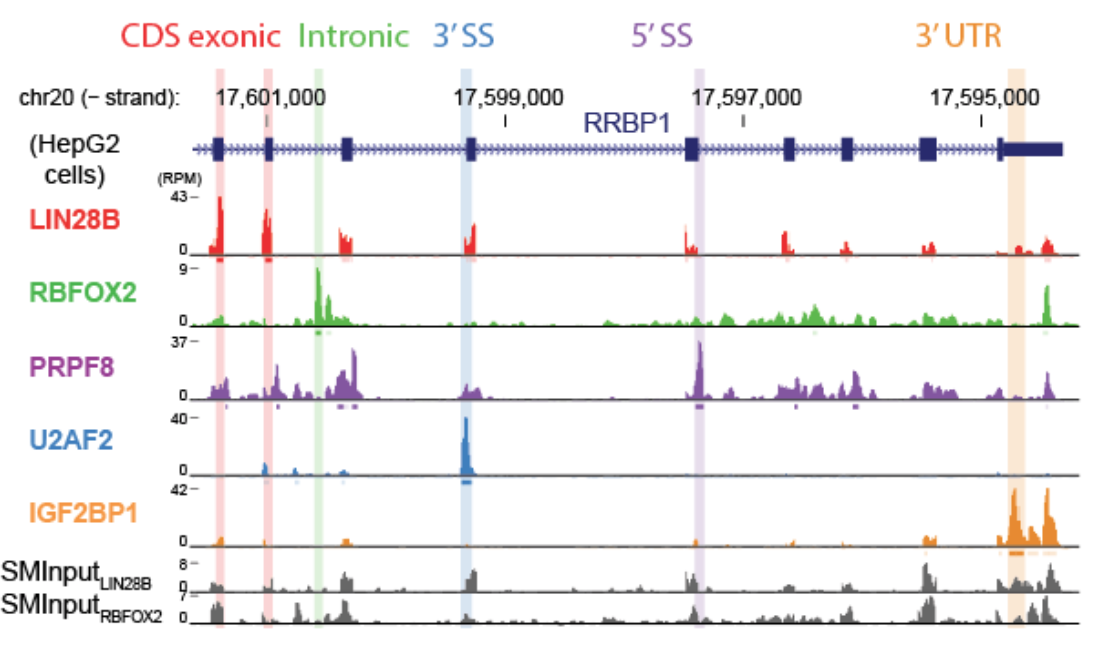
eCLIP-seq identifies binding sites throughout RNAs, including binding to intronic and exonic positions of coding sequences, 3′ and 5′ untranslated regions, non-coding RNAs including lincRNAs and microRNAs, retrotransposons and other RNA transcripts. See above: Example binding profiles for 5 RBPs
- Single-nucleotide resolution
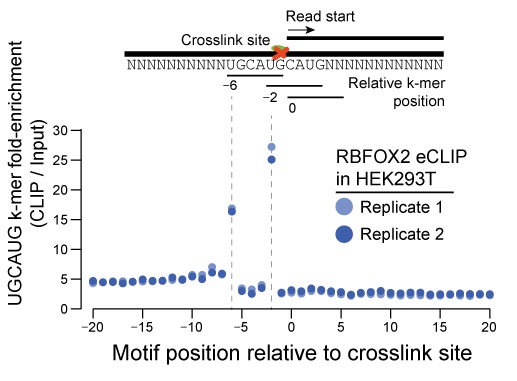
Enhanced CLIP-Seq
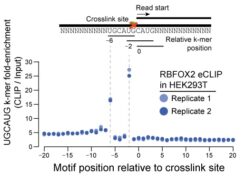
Reverse transcription often terminates at the protein-RNA crosslink site. By performing adapter ligation at the cDNA step, eCLIP can be used to identify binding sites and binding motifs with single-nucleotide resolution, depending on the target protein. See above: Crosslink-termination analysis on RBFOX2
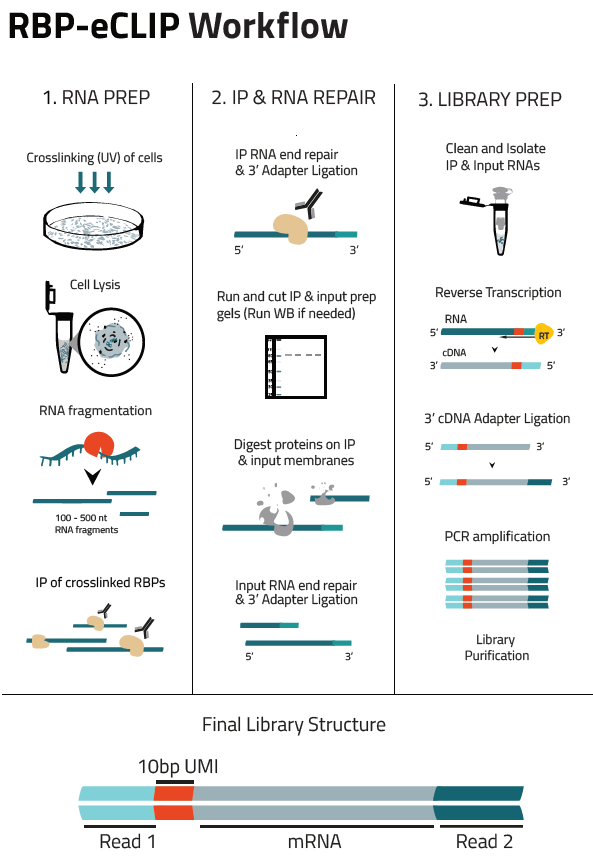
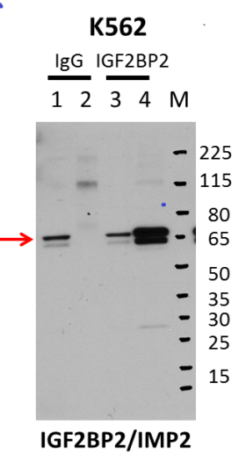
RBP-RNA interactions are covalently linked using UV crosslinking of live cells. Cells are then lysed, and RNA is fragmented using limited RNase treatment. A specific RBP (and its bound RNA) is then immunoprecipitated using an antibody that specifically recognizes the targeted RBP. After ligation of a 3’ RNA adapter, immunoprecipitated material (as well as a paired input sample) are run on denaturing protein gels and transferred to nitrocellulose membranes. A region from the protein size to 75 kDa above is cut from the membrane and treated with Proteinase K to release RNA. After cleanup, RNA is then reverse transcribed to ssDNA, after which a second adapter is ligated. PCR amplification is then used to obtain sufficient material for high-throughput sequencing.
Showing all 2 results