SARS-CoV-2 and Influenza/RSV NGS Amplicon Panels
Nucleus Biotech are proud to cooperate with Paragon Genomics who have developed amplicon panel based NGS kits to support SARS-CoV-2, Influenza and RSV research.
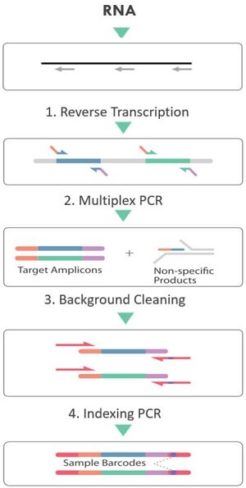
All respiratory research panels from Paragon Genomics are based on their proprietary CleanPlex technology which is superior to other amplicon panels or approaches such as hybridization capture based enrichment or metagenomic sequencing. The technology inherits major advantages associated with traditional multiplex PCR-based methods while overcoming shortcomings such as PCR background noise, scalability (panel size), uniformity and limitation on detecting novel mutations.
 CleanPlex SARS-CoV-2 Flex Panel
CleanPlex SARS-CoV-2 Flex Panel
- Whole SARS-CoV-2 Genome Sequencing from RNA samples of Covid-19 positive patients even with very low viral loads
- Degenerated PCR Primers, strategically designed for detection of new mutations incl. N501Y, del 69H/V70, L452R, P681R etc.
- CleanPlex Technology– advanced PCR primer and background cleaning chemistry for extremely accurate variant calling at less sequencing costs
- In Process Control: Human mRNA primers are included as library preparation control
The CleanPlex SARS-CoV-2 FLEX Panel kit is based on the original CleanPlex SARS-CoV-2 Panel developed early in 2020 as response to the COVID-19 pandemic. The panel enables SARS-COV-2 research and surveillance by sequencing the whole virus genome from samples with even very low SARS-CoV-2 viral content . Panel design is based on the SARS-CoV-2 sequence NC_045512.2. Primers were optimized to preferentially amplify the SARS-CoV-2 cDNA versus the background human cDNA or DNA and uniformly amplify the covered genome. The expertly designed panel allows for the interrogation of the entire viral sequence with as little as 200,000 sequencing reads per samples.
The CleanPlex SARS-CoV-2 FLEX Panel kit features additional sets of degenerated primers for even coverage even when the virus mutates. Thus the FLEX panel offers sensitive detection and robust characterisation of the whole SARS-CoV-2 virus genome for mutation analysis, wastewater surveillance and public health research. To fully cover and reliably identify SARS-CoV-2 variants, Paragon Genomics developed the SARS-CoV-2 Emerging Variants Add-on primer pool which operates as an easy spike-in panel to maintain even coverage and confident identification of the defining mutations of variants such as Omicron including sub-lineages BA.1, BA.2, BA.2.12.1, BA.2.75, BA.3, BA.4 and BA.5, Delta variant B.1.617.2, as well as Alpha V1, Beta V2, Gamma V3, and Mu. The Add-on pool comes with all SARS-CoV-2 Panels free of charge.
SARS-CoV-2 Host NGS Panel
Research has shown that the SARS-CoV-2 virus is internalized via binding of its Spike Protein to the host´s ACE2 with the help of TMPRSS2 expressed by the host cells. The binding affinity of SARS-CoV-2 with ACE2, along with ACE2 and TMPRSS2 expression levels, are major determinants of viral replication rate and disease severity. The CleanPlex ACE2 & TMPRSS2 NGS panel targets all exonic and promoter regions of the two genes and facilitates further investigations in host genome mutations related to the ACE2 binding affinity and expression of ACE2 and TMPRSS2, providing additional insights into the individual’s viral susceptibility and disease’s severity.
 CleanPlex® Respiratory Virus Research Panel
CleanPlex® Respiratory Virus Research Panel - Strategically Designed Content: RSV and Influenza subtype detection and complete SARS-CoV-2 genome sequencing.
- Ultra-sensitive Detection: Multiplex RT-PCR based amplification for sensitive detection
- Fast and Streamlined Workflow: Generate sequencing-ready libraries in just 6 hours using a rapid, four-step protocol from extracted RNA to sequence ready libraries
There is an emerging need for assaying common respiratory viruses concurrently. The CleanPlex Respiratory Virus Research NGS Panel Kit combines SARS-CoV-2 whole-genome sequencing with expertly designed primers specific to influenza A subtypes H1N1, H1N2, H3N2, influenza B, and respiratory syncytial virus (RSV) A&B. The kit detects multiple viral pathogens to not only allow for differentiation of the respiratory tract viruses but also enables sub-typing and strain information for mutation analysis. By spiking in the SARS-CoV-2 Emerging Variants Add-on primer pool the newest SARS-CoV-2 variants such as Omicron including sub-lineages BA.1, BA.2, BA.2.12.1, BA.2.75, BA.3, BA.4 and BA.5 are evenly covered and identified. The Add-on pool comes with all Respiratory Virus Research Panels free of charge.
The Respiratory Virus Research Panel is aiding early warning wastewater surveillance programs to monitor viruses like SARS-CoV-2, Influenza and RSV and enables researchers to discover new vaccine-resistant virus variants.
Showing all 12 results