miRNA Detection
BioLiqX HS miRNA qRT-PCR Kits for High Sensitivity miRNA Detection
• Pre-amplification step and negation of dilution effect ensure superior sensitivity
• Perfect for low copy number circulating miRNAs in biological fluids, such as plasma
• High specificity for any miRNA(s) of your choice; premade kit for organ and mestasis specific miRNAs
• Ideal for validating Small RNA-Seq results quantitatively
• Based on CATL technology: BioLiqX Small RNA-Seq results can be validated directly on the same libraries using BioLiqX HS miRNA qRT-PCR Assays
Workflow of the BioLiqX HS miRNA qRT-PCR Assays
The kits utilize a single-tube protocol for the conversion of all short RNA molecules into a preamplified cDNA library by adding certain reagents and enzymes to the RNA sample in a sequential manner. Subsequently, the preamplified cDNA serves as a template for ultra-sensitive and low-background real-time PCR reactions with Green DNA Dye (a SYBR® Green analog) and miRNA-specific primers. High specificity and sensitivity of BioLiqX HS miRNA Assays are achieved by the optimal concentration of nucleotides, mineral salts, Green DNA Dye and Hot Start Taq DNA Polymerase in the final qPCR reaction. All reactions can be set up at room temperature without the risk of non-specific amplification. The BioLiqX HS miRNA Assays are ideal for detecting low copy number circulating miRNAs in biological fluids but can be also applied to any other sample. The whole procedure can be completed within approximately 8 hours and requires typically a hands-on time between 30-60 minutes depending on the number of samples.
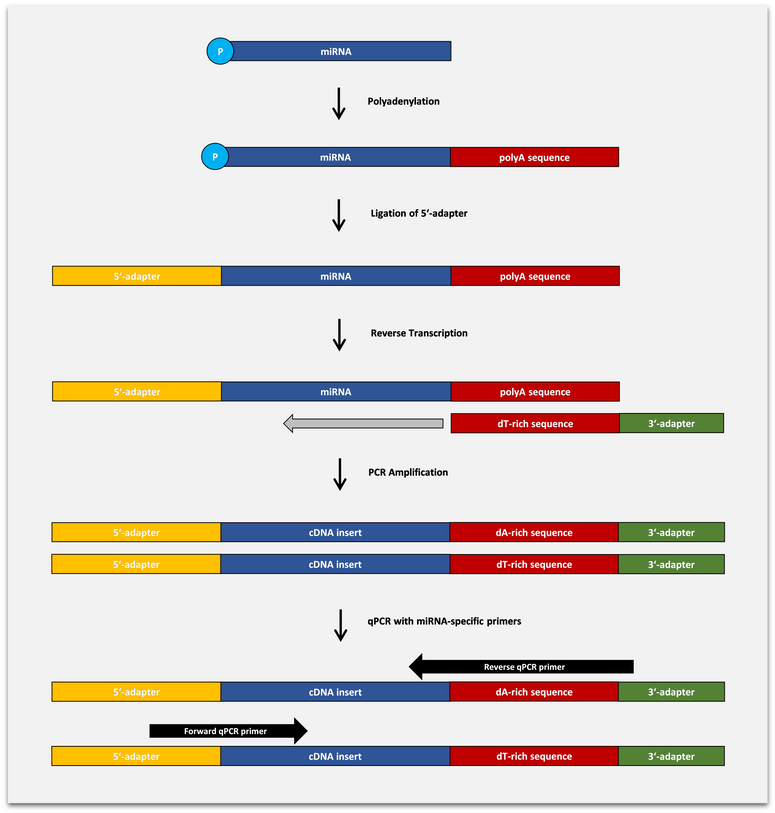
The BioLiqX HS miRNA Assays are based on the so-called “Capture and Amplification by Tailing and Ligation (CATL)” approach for cDNA synthesis and pre-amplification of short or fragmented RNA in the original sample before the final Real-Time PCR step. miRNA is subjected to a polyadenylation reaction followed by the ligation of 5’-adapter. The input RNA flanked by 5’-adapter and 3’-poly(A) tails is then converted into cDNA using anchored RT primer carrying poly(T)-rich sequence and custom 3’-adapter sequence. The cDNA is then PCR amplified (typically for 10-14 cycles) using primers carrying terminal sequences matching the 5′- and 3′-adapters. Finally, the pre-amplified cDNA is used as input for Real-Time PCR reaction with miRNA-specific forward and reverse primers. The Green DNA Dye (a SYBR® Green analog) is used for DNA quantification during Real-Time PCR.
The BioLiqX HS miRNA Assays allow detection of very short cDNA fragments including truncated microRNAs and isomiRs. In addition, pre-amplified libraries generated by the protocol can be used for the simultaneous qPCR analysis of various other cell-free RNA fragments using custom primer pairs.
Superior Assay Performance of BioLiqX HS miRNA qRT-PCR Assays:
BioLiqX HS miRNA Assays have superior sensitivity compared to preamplification-free protocols mainly due to negating RNA and cDNA dilution effect during RT-qPCR and due to the implemented amplification step. The figure below shows the superior sensitivity of the BioLiqX HS miRNA hsa-miR-16, hsa-miR-21 and cel-miR-39 qRT-PCR assays compared to the corresponding preamplification-free protocols from two competing research kits:
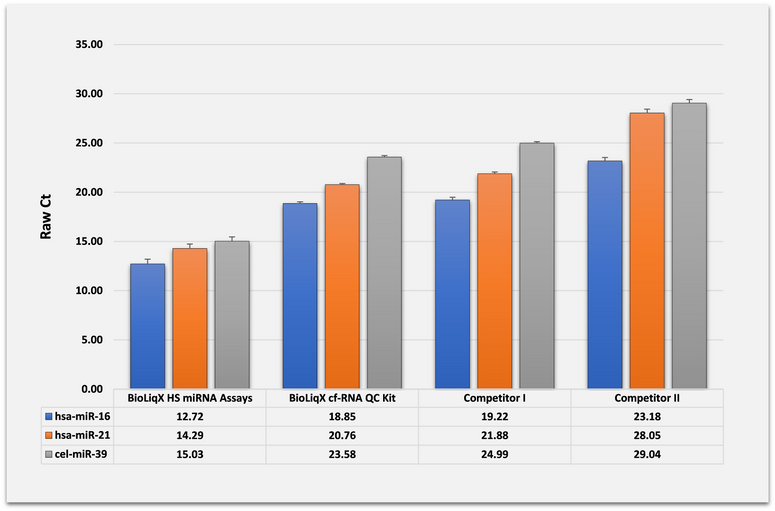
Comparison of Assay Performance:
BioLiqX HS miRNA Assays have superior sensitivity as compared to preamplification-free protocols mainly due to negating RNA and cDNA dilution effect during RT-qPCR. The figure shows the benchmarking of BioLiqX HS miRNA hsa-miR-16, hsa-miR-21 and cel-miR-39 assays with the corresponding preamplification-free assays from either BioLiqX cf-RNA Isolation QC kit or two competing research kits.
Specifically, total RNA was isolated from 0.4 mL of human blood plasma with 0.5 pg 22 nt cel-miR-39 control premixed after the initial lysis. The RNA was eluted in the total volume of 50 µL, and 5 µL of eluates were taken as inputs for each reaction (in triplicates). The BioLiqX HS miRNA libraries were subject to 12x pre-amplification cycles and diluted 10-fold in nuclease free water. The cDNA generated by BioLiqX cf-RNA Isolation QC kit and the competing kits was diluted 6-fold. The proportions of the diluted cDNA in the final qPCR reactions were equal for each protocol. The Real-Time PCR was performed using LightCycler 480 Real-Time PCR System (Roche) according to the recommendations of each kits’ manuals. Raw threshold cycle (Ct) values were determined by the second derivative max method. Each bar represents the mean (SD) of three replicates.
Showing all 2 results

