miRNA Target Sequencing
MicroRNA Target Identification by direct miR-eCLIP sequencing of miRNA-mRNA interactions
• High Resolution Maps of the miRNA-mRNA Interactome
• Direct Detection of mRNA targets
• Reveal functional miRNA targets
• Optional enrichment of miRNAs, miRNA families, or genes for distinct target profiling
• Service also applicable to look for siRNA off-target sites
Precise microRNA (miRNA) target identification is essential to understand post-transcriptional gene regulation. With more than 2000 miRNAs in the human genome, miRNA regulation has been shown to be involved in nearly every physiological system and miRNA misregulation is linked to human diseases; however, identification of the mRNA targets of miRNAs remains a critical challenge.
miR-eCLIP™ enables precise mapping of direct miRNA-mRNA interactions transcriptome-wide and can also be utilized to look at off-targets of siRNA. In contrast to alternative techniques that provide indirect methods to map miRNA targets (i.e., computational algorithms or over-expression/knockdown experiments), miR-eCLIP identifies direct in vivo miRNA targets using AGO2 immunoprecipitation, RNA-RNA ligation, and high-throughput sequencing (similar to methods such as CLASH or CLEAR-CLIP). Further, miR-eCLIP has the option of enriching for specific miRNAs or genes of interest, enabling profiling of miRNA-target interactions at an unprecedented depth.
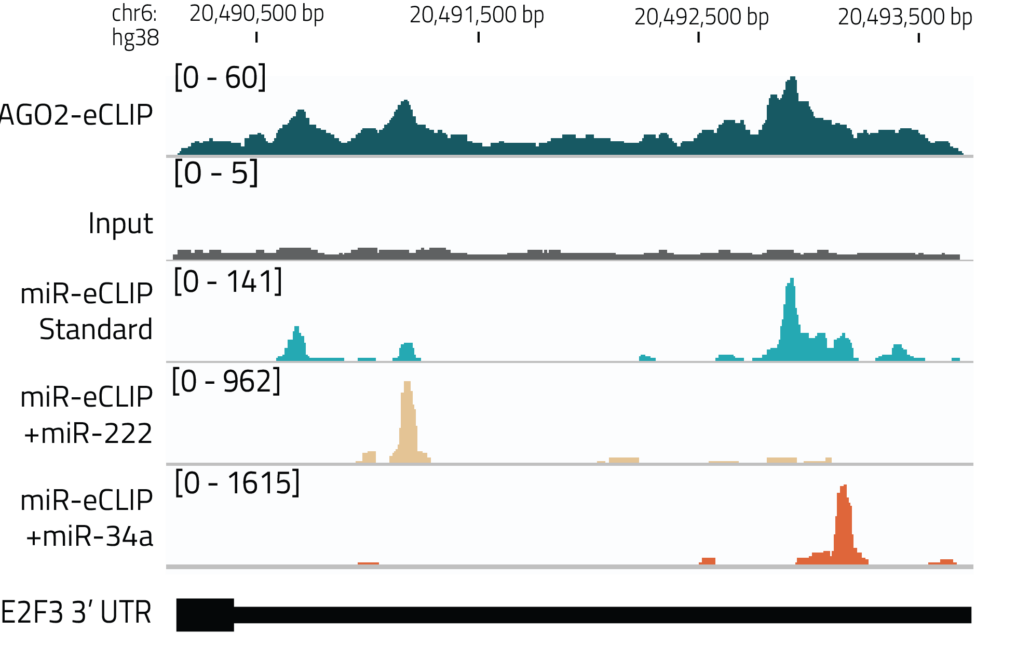
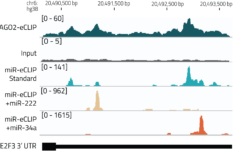
Direct detection of miRNA targets by sequencing:
miR-eCLIP uses a modified AGO2-eCLIP protocol to enhance ligation of miRNAs to target mRNAs yielding chimeric miRNA-mRNA fragments. Thousands of chimeric miRNA-mRNA reads are utilized for direct and functional mRNA target identification.
Showing the single result