NGS Reagents for Pooled shRNA Libraries
 NGS Reagents for Pooled shRNA Libraries are tools to amplify and sequence shRNAs, after running screens with pooled lentiviral libraries.
NGS Reagents for Pooled shRNA Libraries are tools to amplify and sequence shRNAs, after running screens with pooled lentiviral libraries.
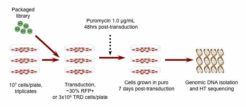
For RNAi screens, the frequency of each hairpin is assessed by next-generation sequencing (NGS) to determined which ones are enriched or depleted relative to the original libraries or control samples.Nucleus Biotech provides NGS Prep Kits for various libraries to meet your needs. Each contains the appropriate primers for particular library/vector combinations to generate sequencing-ready samples. Simply select the Prep Kit for your screen and order.
Showing all 4 results