RNA Methylation Sequencing: Enhanced m6A-seq/meRIP-seq
High Resolution Maps of the Epitranscriptome – Efficient and precise identification of m6A modification sites
N6-Methyladenosine (m6A) is the most abundant modified nucleoside in mRNA. The dynamic epitranscriptomic m6A RNA modification by writers and erasers and its recognition by readers is regulating cellular and physiological processes and is involved in many diseases.
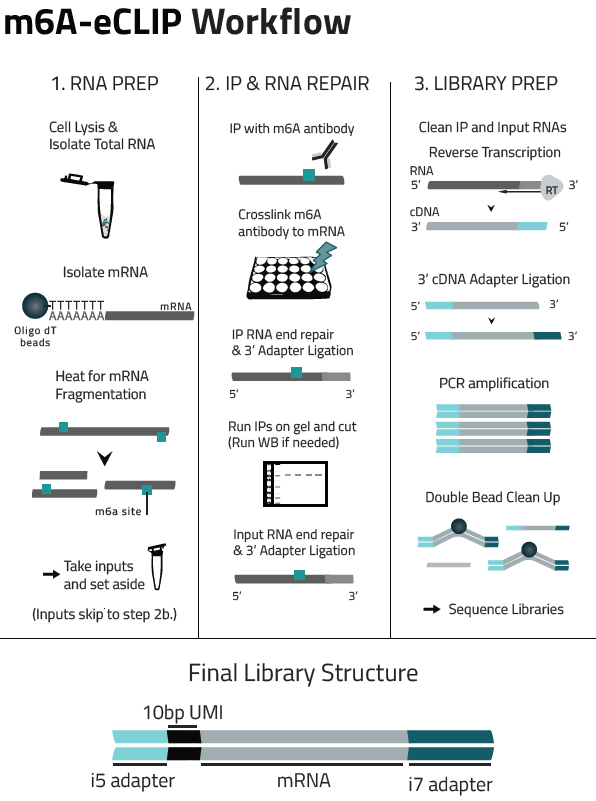
Eclipse BioInnovation´s m6A-eCLIP-seq Kit is an NGS-based assay for the most sensitive, accurate and reproducible detection of m6A modification sites, superior to meRIP-seq protocols: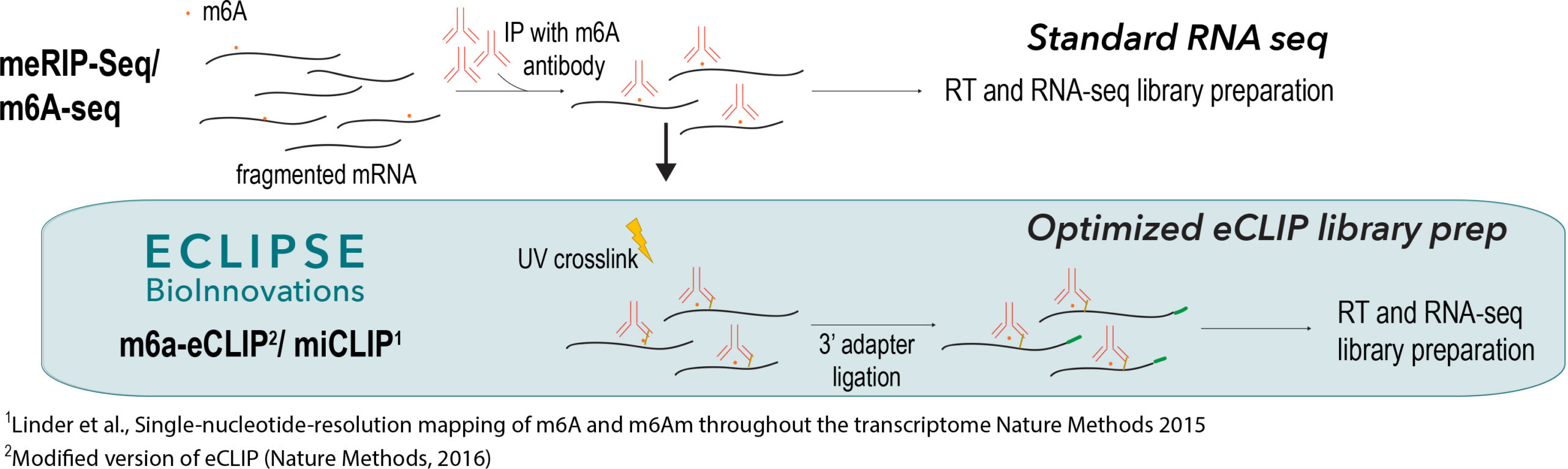
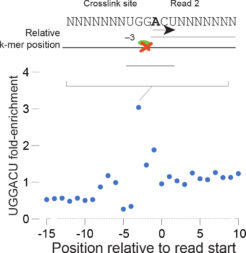
⇒ Accurate Single Nucleotide Resolution
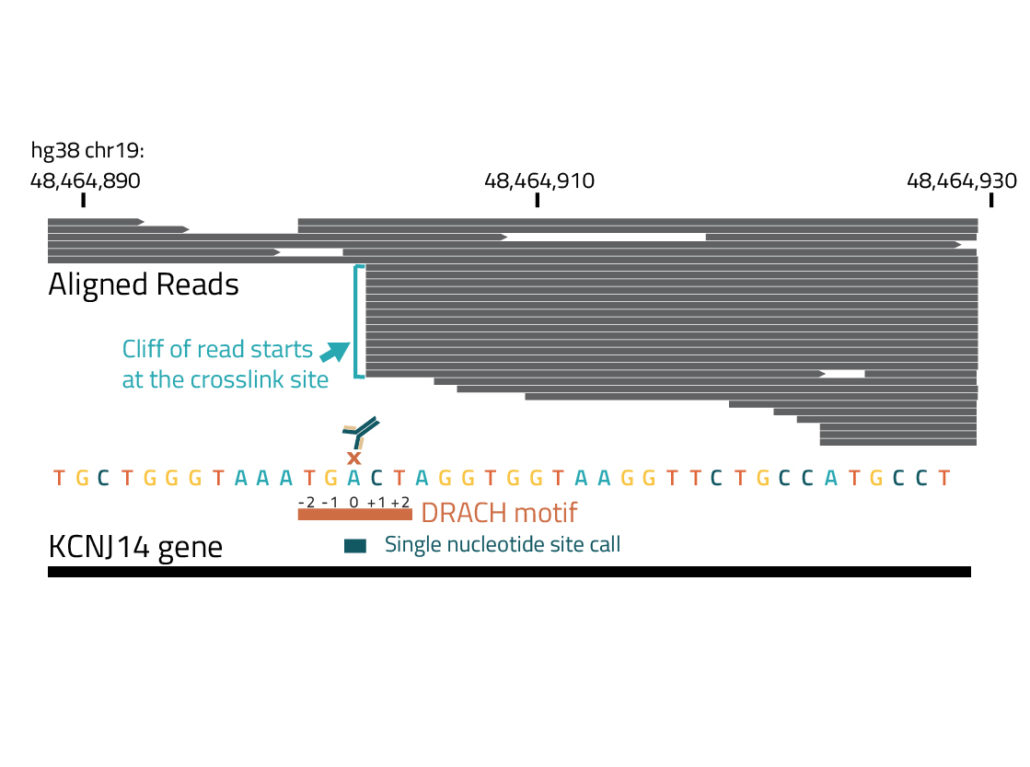 Reverse transcriptase (RT) termination at the crosslink sites of the m6A antibody to RNA form sharp cliffs of aligned read starts enabling single-nucleotide resolution modification calls. More than 70% of identified sites are positioned on adenosine nucleotides, and over 50% of all crosslink sites map precisely to adenosines in the DRACH motif preferred by m6A methyltransferases
Reverse transcriptase (RT) termination at the crosslink sites of the m6A antibody to RNA form sharp cliffs of aligned read starts enabling single-nucleotide resolution modification calls. More than 70% of identified sites are positioned on adenosine nucleotides, and over 50% of all crosslink sites map precisely to adenosines in the DRACH motif preferred by m6A methyltransferases
Showing the single result

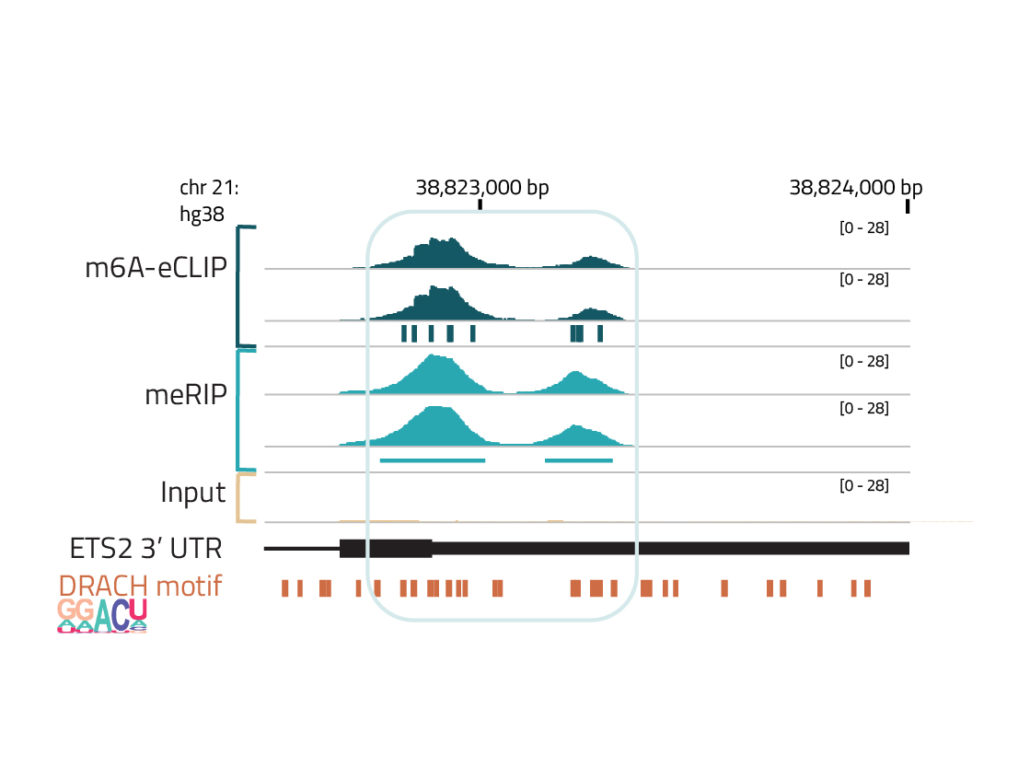 Consistent with the known feature of m6A modifications, peaks of m6A-eCLIP reads are enriched near the stop codons, and for the 5 nt DRACH motif. m6A-eCLIP shows similar read densities to a low-resolution m6A profiling technique m6A-RIP (meRIP), but with more precise site calls, and requiring 100-fold less mRNA input.
Consistent with the known feature of m6A modifications, peaks of m6A-eCLIP reads are enriched near the stop codons, and for the 5 nt DRACH motif. m6A-eCLIP shows similar read densities to a low-resolution m6A profiling technique m6A-RIP (meRIP), but with more precise site calls, and requiring 100-fold less mRNA input.