Small RNA-Seq Kits
BioLiqX Low Input Small RNA–Seq Kit: High Efficiency Analysis of cf-RNA
- Ultra high sensitivity – Input of as little as 1 pg small RNA
- Easy automation friendly workflow – Half day protocol
- Uniform transcript representation – Equivalent detection of miRNA as well as fragmented mRNA and lncRNA
- No sequence bias – Even transcript length and GC content distribution
- Highly reproducible sequencing results
- Bioinformatics services optionally offered
BioLiqX Small RNA-Seq Kit:
CATL Technology enables ultra low inputs of as little as 1 pg
Nucleus Biotech is pleased to present the BioLiqX Low Input Small RNA-Seq Kit, developed by Heidelberg Biolabs for liquid biopsy samples such as plasma, serum, and EVs (Extracellular Vesicles, e.g. exosomes), which contain extremely low amounts of cell-free circulating small RNA. It can be also used for other inputs including fragmented cellular total RNA or enriched small RNA fractions of cellular RNAs.
The BioLiqX Low Input Small RNA-Seq Kit employs CATL (Capture and Amplification by Tailing and Ligation) – an advanced know-how technology developed by a group of scientists from German Cancer Research Center who had previously explained the origin and high stability of extracellular circulating microRNAs in biofluids (Turchinovich, A., Weiz, L., Langheinz, A. & Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res 39, 7223-7233 (2011). Turchinovich, A. & Burwinkel, B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol 9, 1066-1075 (2012). In total, developers of Heidelberg Biolabs products have more than 15 years’ research experience in the field of circulating nucleic acids, including methods development for high-throughput sequencing and RT-qPCR. Their work resulted in multiple highly cited original research reports and reviews: https://www.heidelbergbiolabs.de/resources/our-publications.html
CATL uses 3´ polyadenylation and 5´ adaptor ligation as well as a subsequent amplification step, resulting in highly efficient unbiased NGS library preparation:
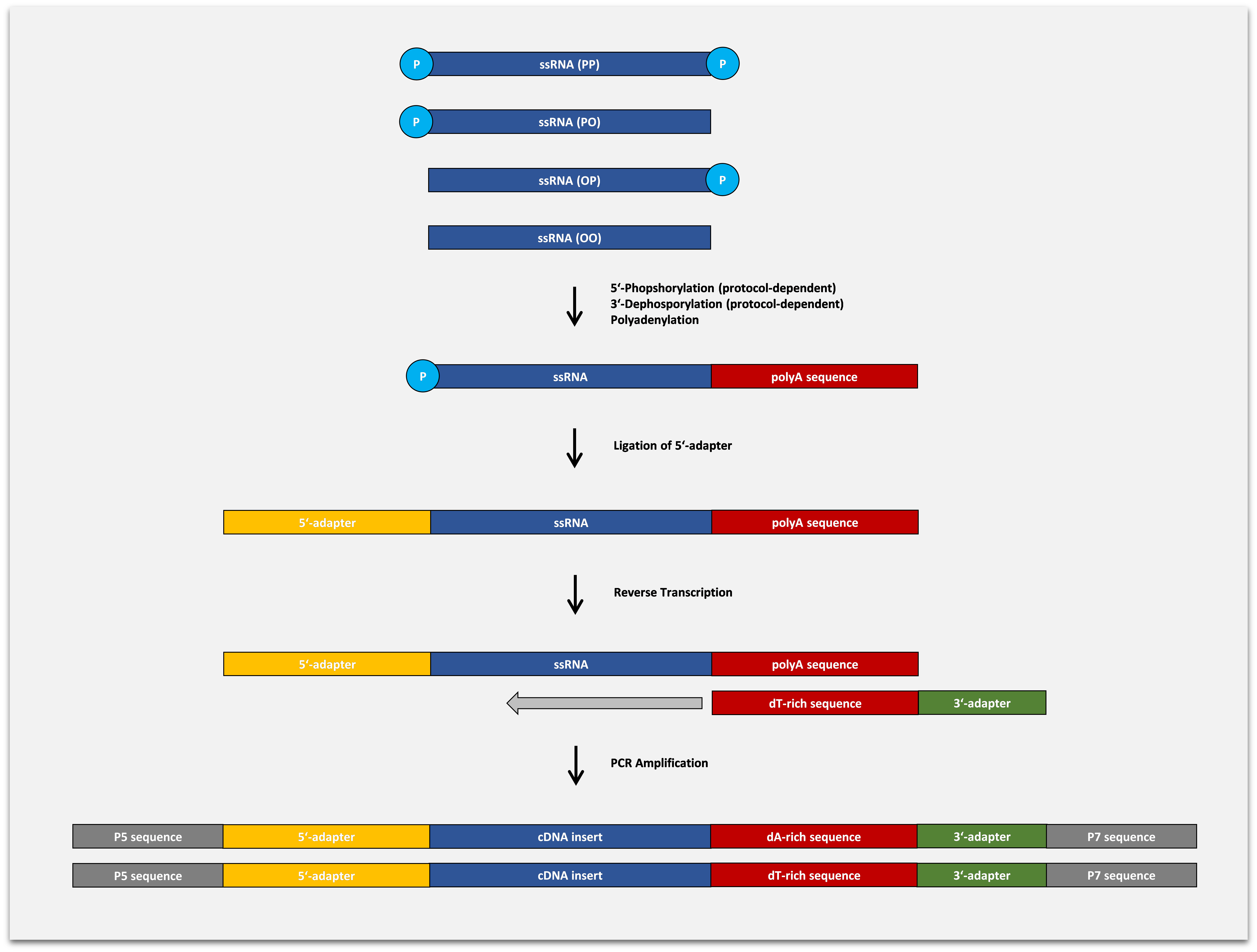
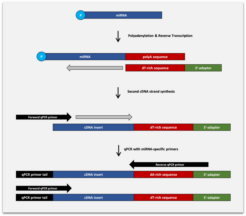
Capture and Amplification by Tailing and Ligation (CATL) Technology for Small RNA-Seq
The BioLiqX Small RNA-seq Kit is based on the Capture and Amplification by Tailing and Ligation (CATL) approach to generate sequencing libraries for Illumina platforms:
Fragmented single-stranded RNA molecules undergo polyadenylation at the 3′-termini and adapter ligation at the 5′-termini. In the follow-up step, the RNA is converted into cDNA by reverse transcription in the presence of the anchored RT primer containing a sequence complementary to the polynucleotide tail and a custom 3′-terminal sequence. The second cDNA strand is generated during the first cycle of the final pre-amplifying PCR reaction. Multiplexing i7 indexes are introduced during the final PCR amplification with primers carrying P5 and P7 terminal sequences required for cluster generation on Illumina machines.
Polynucleotide tailing is a significantly more efficient way to capture 3-OH termini of single-stranded nucleic acids as compared to 3´ adapter ligation and has little or no biases. As a result, DNA libraries produced by the CATL approach have higher complexity as compared to libraries generated by purely ligation-based methods, Besides, CATL allows qPCR detection of single miRNA copies and customized massive parallel sequencing from picogram inputs of RNA.
• Uniform transcript representation – Equivalent detection of miRNA as well as fragmented mRNA and lncRNA
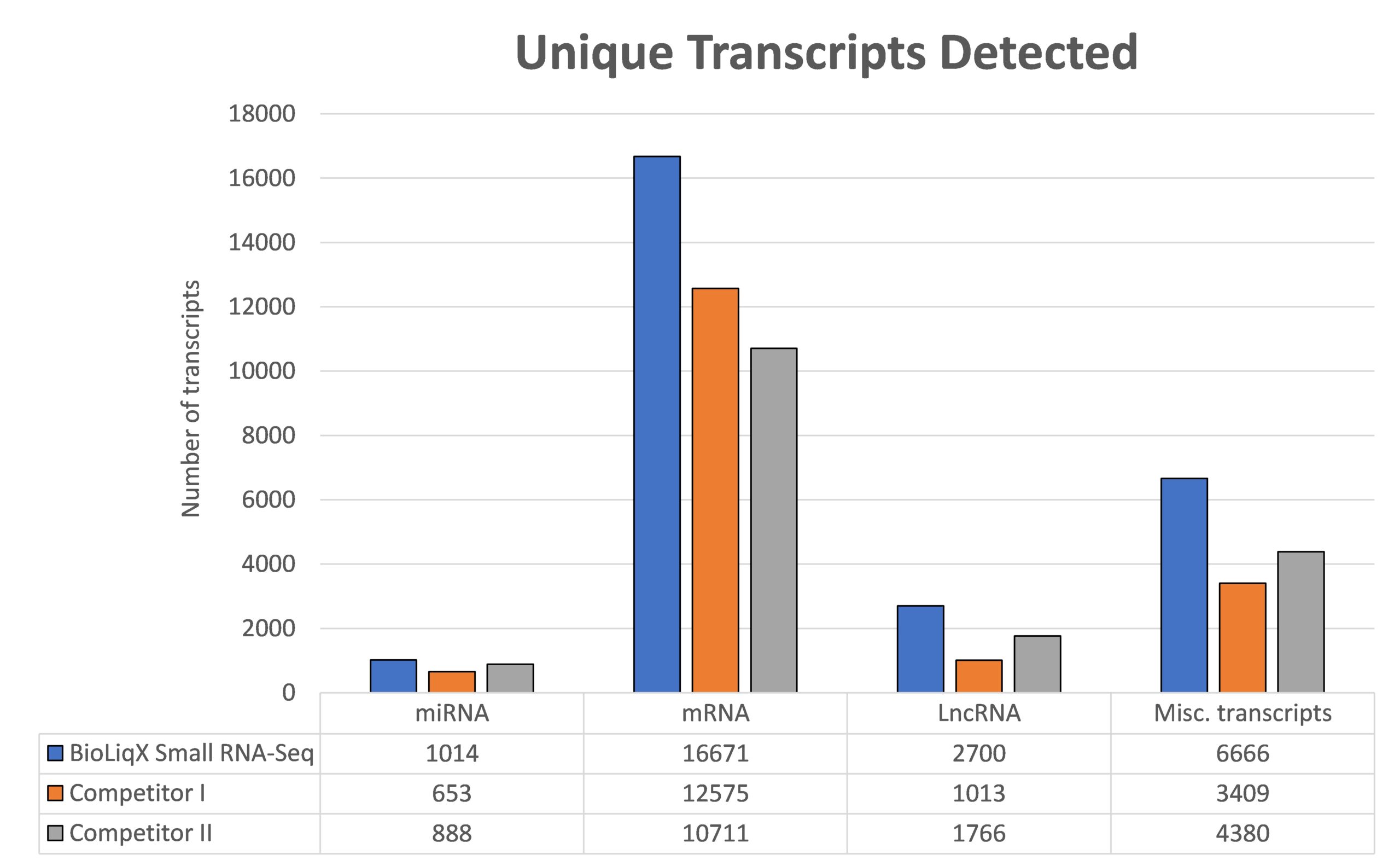
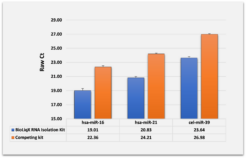
Heidelberg Biolabs´ BioLiqX kit was compared to the two market leading Small RNA-Seq kits using the same input material for all 3 NGS library prep kits (0.5 ml blood plasma): The BioLiqX kit enabled sequencing of the significantly highest variety across all transcript classes – miRNA, mRNA, and lncRNA
• No sequence bias – Even transcript length and GC content distribution
Heidelberg Biolabs´ BioLiqX kit was compared to the two market leading Small RNA-Seq kits using the same input material for all 3 preps: Varying transcript lengths and GC contents were representatively sequenced using the BioLiqX NGS library prep kit in contrast to the other kits which preferentially detected larger RNAs (competitor 1/company C) or shorter RNAs (competitor 2/company Q) and did show uneven GC distribution:
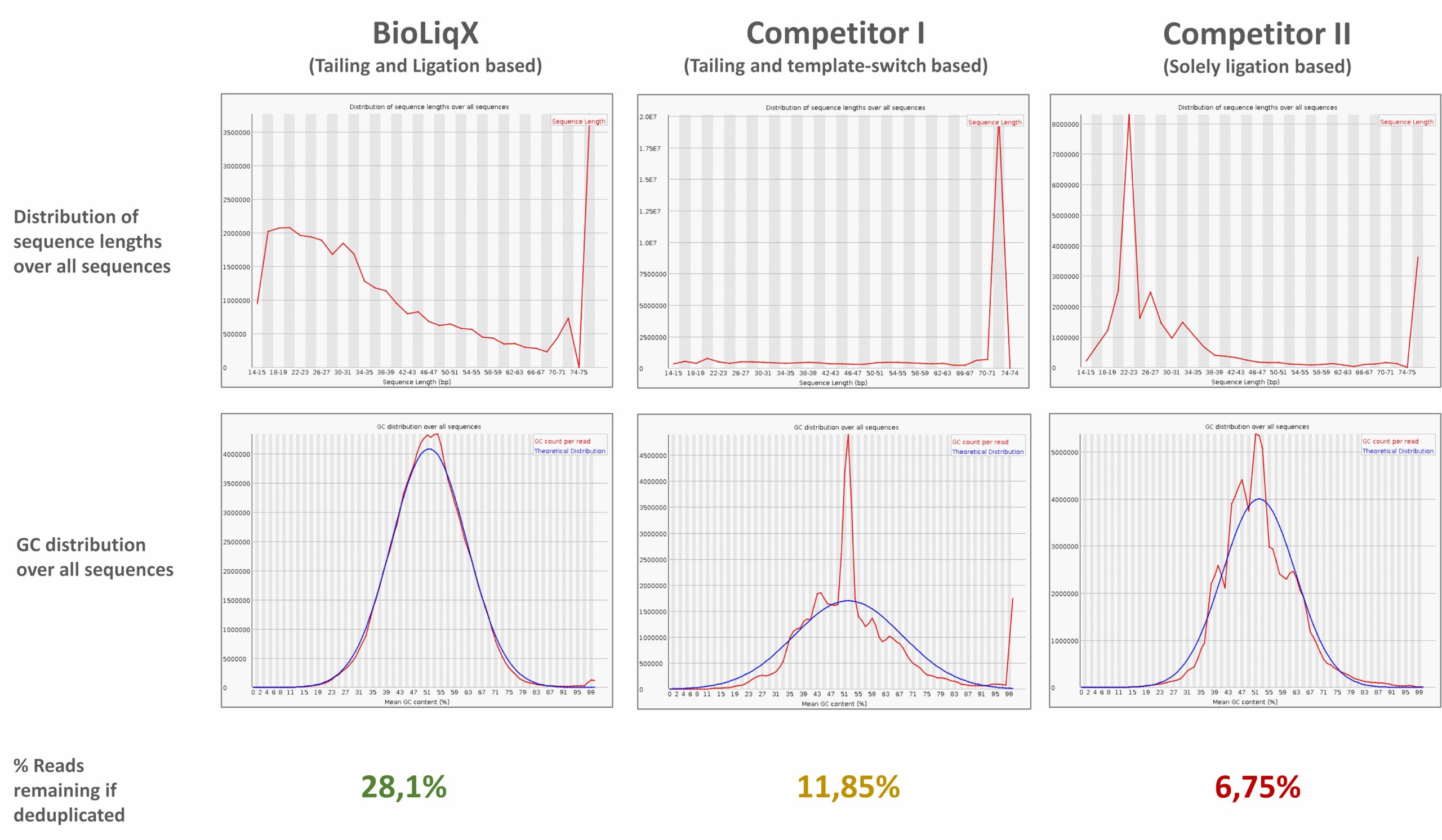
Heidelberg Biolabs´ BioLiqX kit was compared to the two market leading Small RNA-Seq kits using the same input material for all 3 NGS library prep kits (100 pg small cf-RNA from human blood plasma): In contrast to the other methods, the BioLiqX kit employing CATL enabled unbiased sequencing of varying transcript lengths and GC contents
• Highly reproducible sequencing results
After primer removal, BioLiqX small RNA-seq NGS libraries (three independent replicates) were sequenced on the Illumina HiSeq2000 platform. The obtained raw FASTQ files were trimmed from polyA-tails and size-selected using cutadapt software:
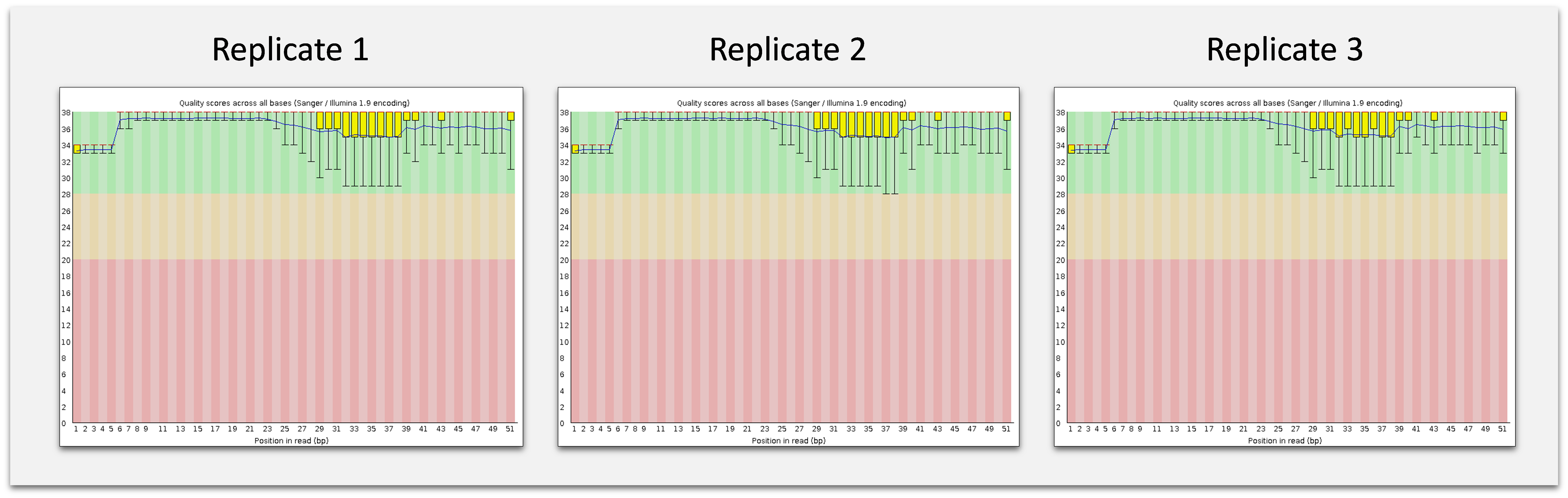
Quality score reports obtained by fastqc software after HiSeq2000 sequencing of three independent BioLiqX Small RNA-seq libraries generated from a human plasma RNA sample. The raw FASTQ files were trimmed using cutadapt. The reads shorter then 15 nt were discarded.
Trimmed and size-selected reads were aligned to the custom curated hg38 reference transcriptomes in a sequential manner. First, all reads were mapped to RNA species with low sequence complexity and/or high number of repeats: rRNA, tRNA, RN7S, snRNA, snoRNA/scaRNA, vault RNA, RNY as well as mitochondrial chromosome (mtRNA). All reads that did not map to the above RNAs were aligned sequentially to mature miRNA, pre-miRNA , protein-coding mRNA transcripts (mRNA) and long non-coding RNAs (lncRNAs). The reads which did not map to the above RNAs were aligned to the remaining transcriptome [other ncRNAs containing mostly pseudogenes and non-protein coding parts of mRNAs]. Finally, all reads which did not map to human transcriptome were aligned to the human genome reference (rest hg38) that corresponds to introns and intergenic regions. The raw count tables from each replicate were combined and compared for reproducibility using correlation plots:
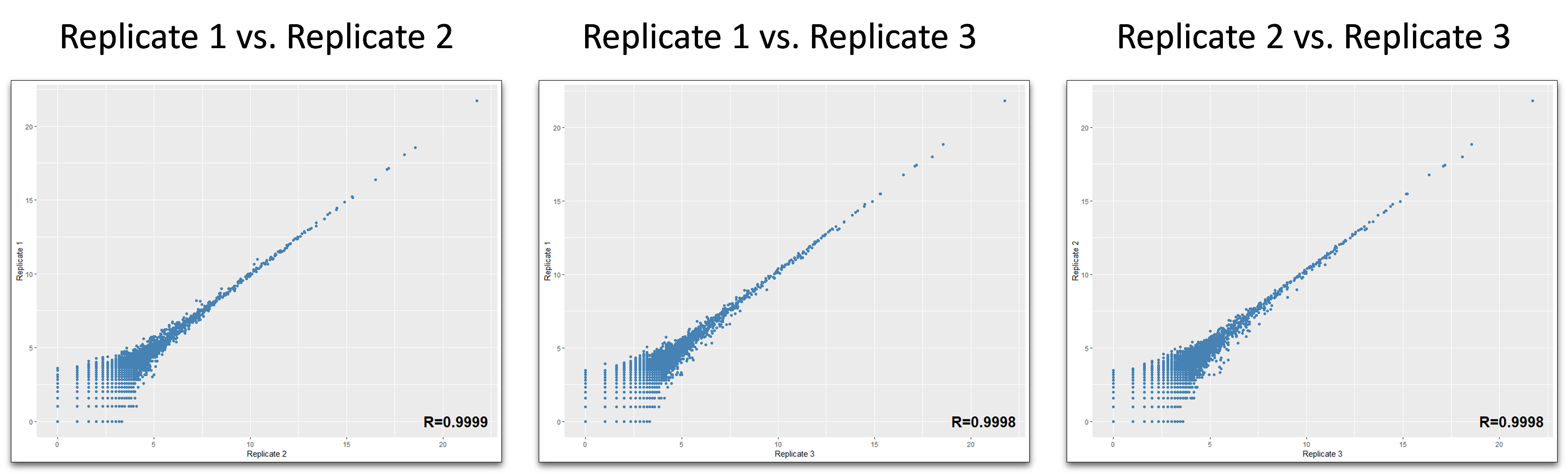
Correlation plots of raw human transcriptome reads obtained from three independent BioLiqX small RNA-seq libraries generated from human plasma RNA sample and sequenced on HiSeq2000
Bioinformatics Services
Fully customized computational analyses of massive parallel sequencing data using validated bioinformatics pipelines tailored for circulating RNA-seq are available for you, when needed – please contact us with your project details
Recommended Products for cf-RNA Isolation and cf-RNA Isolation Quality Check
As cell-free RNA in liquid biopsy samples such as blood plasma is highly diluted, very efficient isolation procedures are required to obtain high quality input material for subsequent molecular analysis by NGS or qRT-PCR.
Our partner Heidelberg Biolabs have developed unique solutions:
• BioLiqX cf-RNA Isolation Kit: Employs phenol/chloroform-based denaturation of contaminants as well as optimized buffers and columns ensuring the yield of highly concentrated ultrapure cf-RNA
• BioLiqX cf-RNA Isolation QC Kit: Contains all reagents and primers required for the detection of four “house-keeping” miRNAs in any RNA sample using qRT-PCR. The kit also contains synthetic cel-miR-39 for spike-in assessment of small cf-RNA isolation between samples as well as normalization of miRNA expression in biological fluids after downstream qRT-PCR.
Recommended High Sensitivity qRT-PCR Kit for quantitative validation of Small RNA-Seq Results
BioLiqX HS miRNA qRT-PCR Assays from Heidelberg Biolabs are also based on Capture and Amplification by Tailing and Ligation (CATL) technology which ensures the same extremely high sensitivity as well as compatibility with the BioLiqX Small-RNA seq libraries
Showing all 3 results