Description
BioLiqX Small RNA–Seq Kit for High Efficiency Analysis of cf-RNA
• Ultra high sensitivity – Input of as little as 1 pg small RNA
• Easy automation friendly workflow – Half day protocol
• Uniform transcript representation – Equivalent detection of miRNA as well as fragmented mRNA and lncRNA
• No sequence bias – Even transcript length and GC content distribution
• Highly reproducible sequencing results
Nucleus Biotech is pleased to present the BioLiqX Small RNA-Seq Kit, developed by Heidelberg Biolabs for liquid biopsy samples such as plasma, serum, and EVs (Extracellular Vesicles, e.g. exosomes), which contain extremely low amounts of cell-free circulating small RNA. It can be also used for other inputs including fragmented cellular total RNA or enriched small RNA fractions of cellular RNAs.
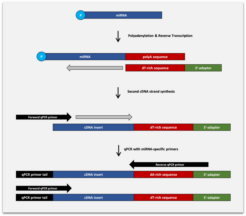
The BioLiqX Small RNA-Seq Kit employs the CATL (Capture and Amplification by Tailing and Ligation) technology, which uses 3´ polyadenylation and 5´ adaptor ligation as well as a subsequent amplification step, resulting in highly efficient unbiased NGS library preparation
Unbiased Small RNA-seq with high sensitivity
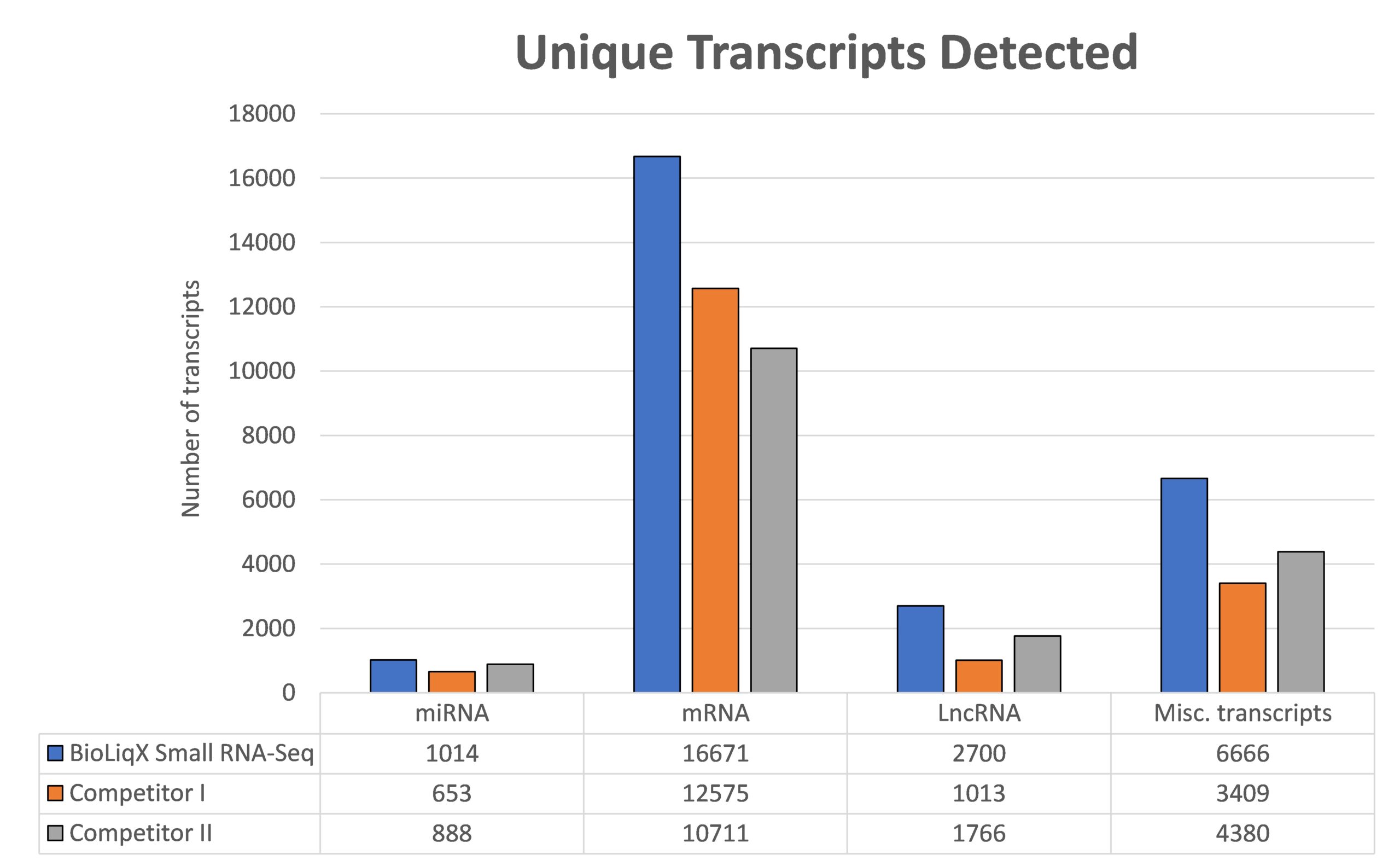
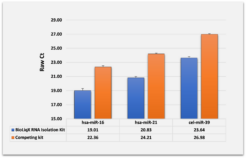
Heidelberg Biolabs´ BioLiqX kit was compared to the two market leading Small RNA-Seq kits using the same input material for all 3 preps: Varying transcript lengths and GC contents were representatively sequenced using the BioLiqX NGS library prep kit in contrast to the other kits which preferentially detected larger RNAs (company C) or shorter RNAs (company Q) and did show uneven GC distribution
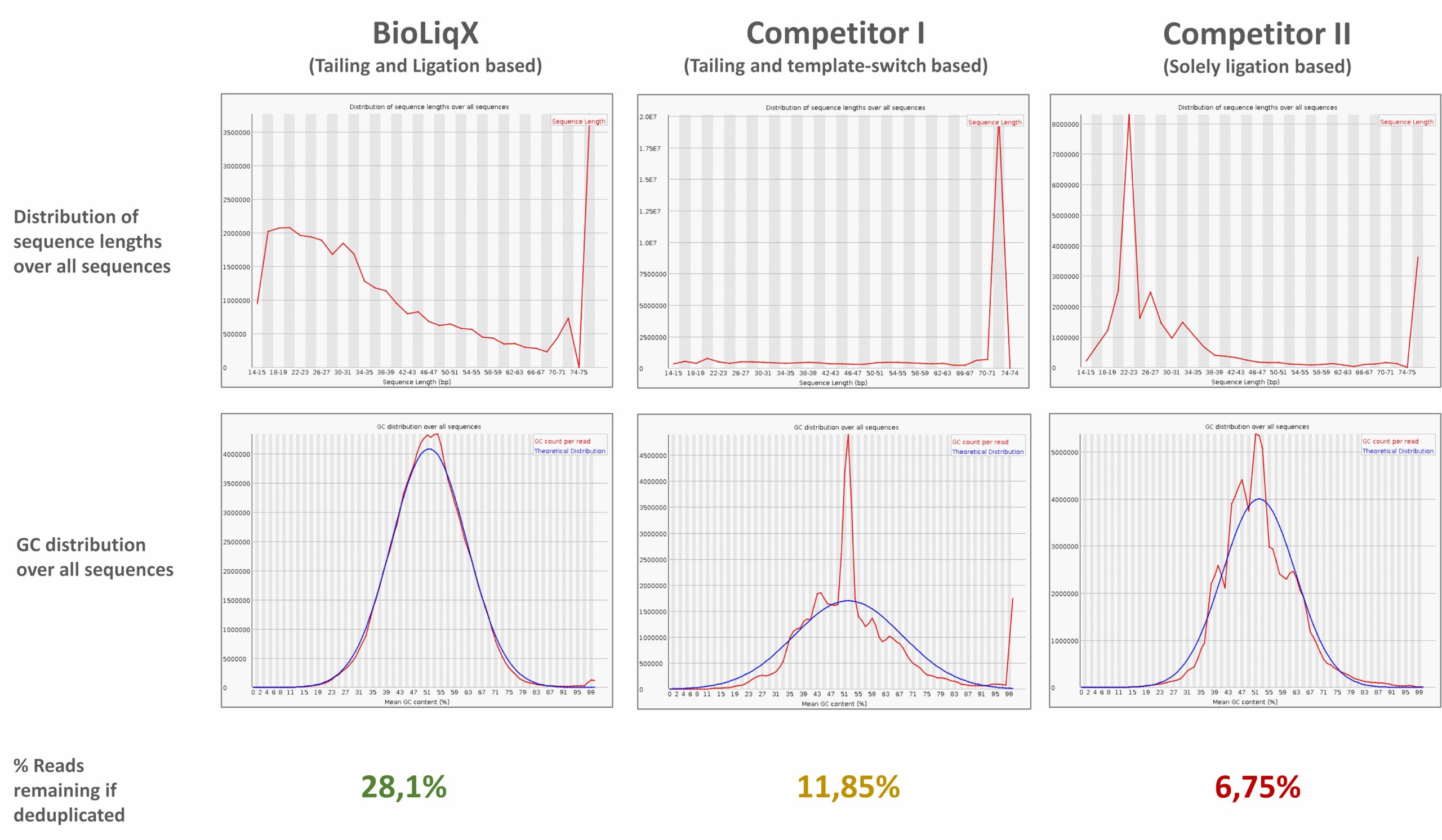
Heidelberg Biolabs´ BioLiqX kit was compared to the two market leading Small RNA-Seq kits using the same input material for all 3 NGS library prep kits (100 pg small cf-RNA from human blood plasma): In contrast to the other methods, the BioLiqX kit employing CATL enabled unbiased sequencing of varying transcript lengths and GC contents
Kit Description
The BioLiqX Small RNA-seq Kit includes all the components required to generate DNA libraries for massive parallel sequencing of small RNA (or long RNA after fragmentation) on Illumina platforms. The kit utilizes a single-tube protocol implying the addition of certain reagents and enzymes to the RNA sample in a sequential manner. The DNA library preparation workflow was originally developed for 1 ng – 1 pg inputs of highly fragmented RNA present in biofluids such as blood plasma and serum (typically between 3-100 nt). However, the kit can be also used for other inputs including total RNA purified from cells or enriched small RNA fractions of cellular RNAs.
Biological fluids (like any other sources) contain distinct populations of RNAs carrying either hydroxyl (-OH) or phosphate (-P) groups at their 5’- and 3’- termini. Those include: (1) 5’- and 3’-phosphorylated RNAs (PP); (2) 5’-OH/3’-OH carrying RNAs (OO); (3) 5’-OH and 3’-phosphorylated RNAs (OP) and (4) 5’-phosphorylated/3’-OH RNAs (PO). For instance, the PO category includes predominantly mature miRNAs and snoRNAs. The BioLiqX small RNA-seq kit was designed to capture RNA carrying distinct 5’- and 3’-terminal groups by including slight modifications of the protocol depending on the task and the scientific question to be addressed. The libraries generated by BioLiqX small RNA-seq kit are stranded, with Read1 and Read2 corresponding to the sense and antisense strand of the input RNA, respectively. The whole procedure can be completed within approximately 6 hours and requires a hands-on time between 20-60 minutes depending on the number of samples.
Kit Contents
The kit includes reagents for polynucleotide tailing and optional end-repair (Tailing Buffer, PdP Enzyme, Tailing Nucleotides, Tailing Enzyme), ligation reagents (Ligation Buffer, Ligation Enzyme), RT Mix and RT primers for reverse transcription, PreAmp Mix for the final cDNA preamplification and a synthetic cel-miR-39 ssRNA control.
By default, the kit is supplied for 24 reactions and includes 24 standard P7 Index primers for multiplexing.
Kit Customization
Any kit sizes larger than 24 rxns can be manufactured for you –
please contact us for a quote!
Dual (P5 and P7) indexed primers can also be included in the kit on request –
please contact us for a quote!