AIRR-seq Spike-in Control Standards
DriverMap AIR RNA Spike-in Controls for Adaptive Immune Receptor Repertoire Sequencing
A collection of synthetic mRNA constructs to be used as universal controls for both commercial and home-brewed AIRR-seq assays based on multiplex RT-PCR or 5’-RACE (SMART) PCR techniques
These first-in-kind RNA spike-in controls are designed to ensure consistent and reproducible T-cell Receptor (TCR) and B-cell Receptor (BCR) clonotype profiling using multiplex RT-PCR such as Cellecta´s DriverMap AIR Assay or 5´-RACE /SMART PCR.
The RNA standards contain nearly full-length V(D)JC structures that represent all seven TCR (TRB, TRA, TRG, TRD) and BCR (IGH, IGK, and IGL) chains:
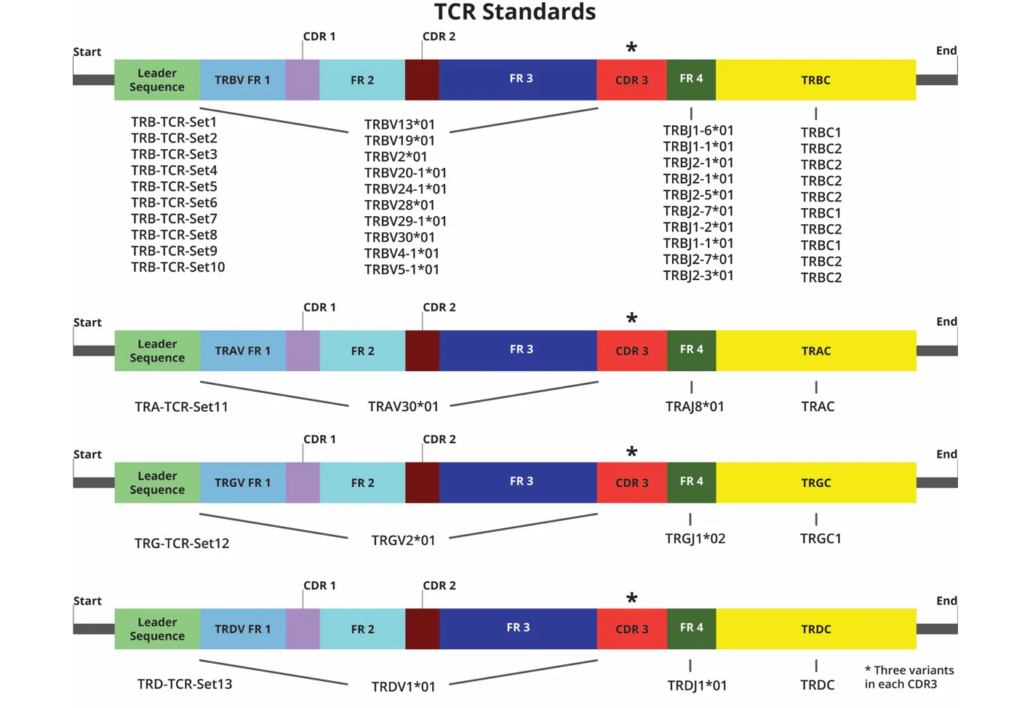
39 TCR constructs, each representing unique TRBs, TRAs, TRGs, and TRDs are shown. Each construct has 3 variations in the CDR3 region (*) that differ by 3 nucleotides in fixed positions
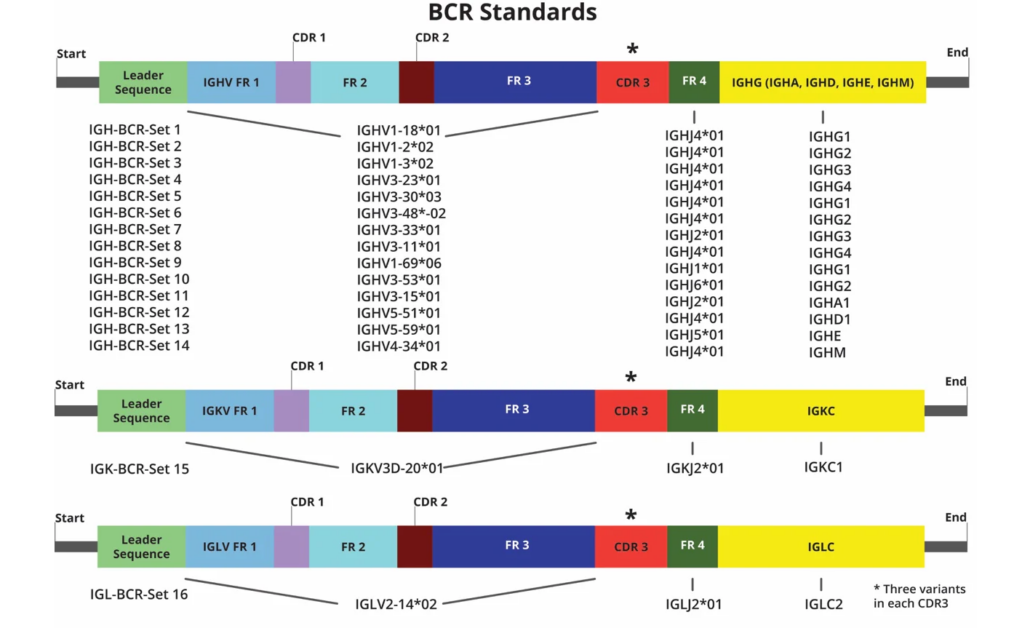
48 BCR mRNA spike-in constructs for IGH, IGK and IGL chains are shown. Each construct has three variations in the CDR3 region (*) that differ by three nucleotides in a fixed position.
Product Formats
DriverMap AIR RNA Spike-in controls are available as Premixed Control or as Sets of Isoform Pools for use in TCR- or BCR-NGS-based AIRR profiling:
- Premixed Control:
- Sets of Isoform Pools:
Applications for DriverMap AIR RNA Spike-in Controls
- Ensure Sensitivity and Specificity: Premixed Control may be spiked into your experimental sample to measure the accuracy and linear range of your PCR assay.
- Error Quantification: Premixed Control may be used to assess sequencing errors introduced at the RT, PCR and NGS stages.
- Sample Quality Control: Isoform Pool Sets or Premixed Control may be used to assess sample quality such as in degraded FFPE samples.
- Cross-Contamination Detection: Isoform Pool Sets can measure cross-contamination between different samples.
Showing all 4 results


