Description
Universal Spike-In Control RNA Mix for commercial and home-brewed AIRR-seq TCR Profiling Assays based on multiplex RT-PCR or 5’-RACE (SMART) PCR techniques
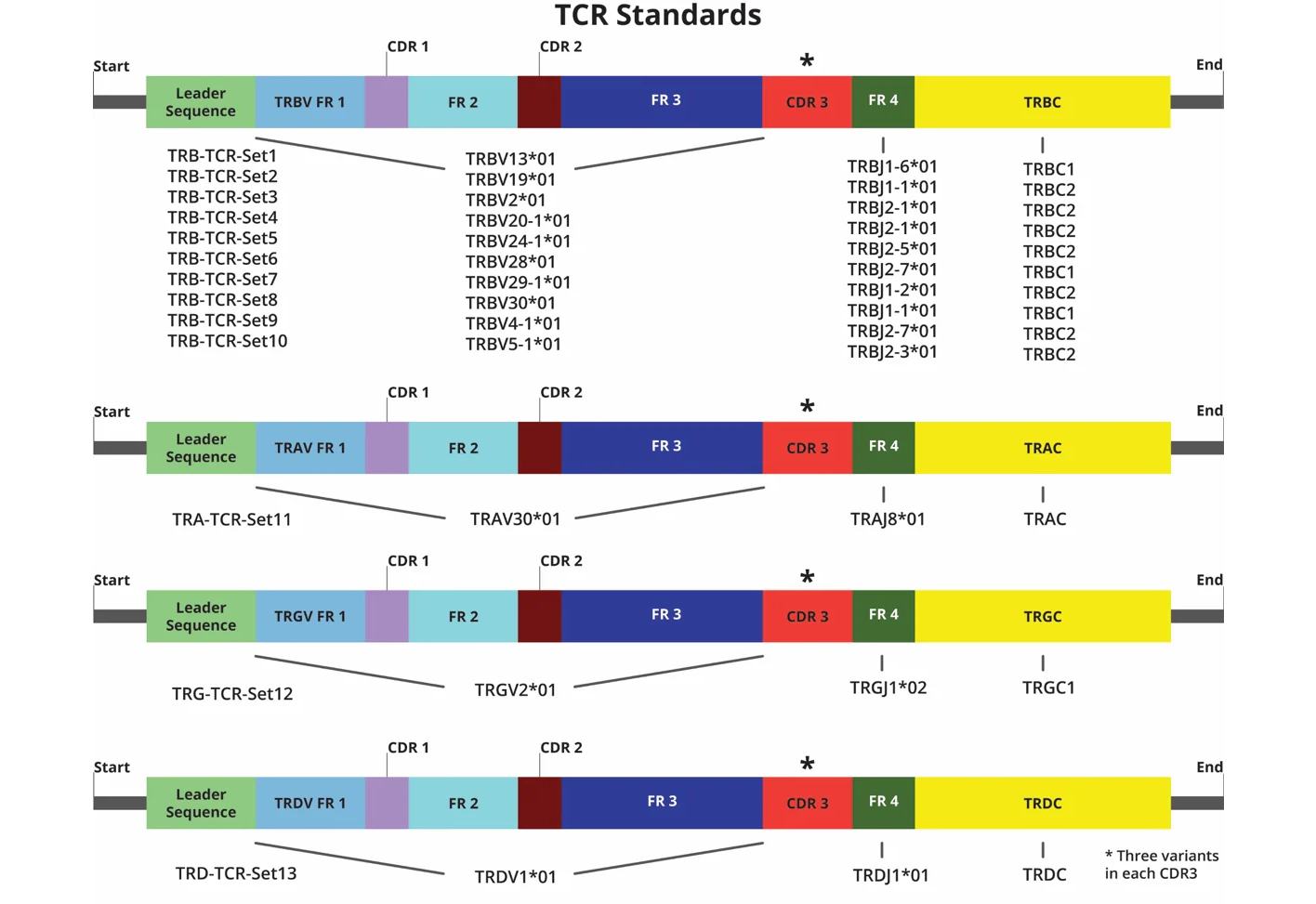
Uniquely available synthetic RNA spike-in control for AIRR-seq, designed to ensure consistent and reproducible T-cell Receptor (TCR) clonotype profiling – contains nearly full-length V(D)JC structures representing all 4 TCR (TRB, TRA, TRG, TRD) chains
The Spike-In RNA Control Mix contains 39 synthetic TCR constructs featuring 13 TCR isoforms with three CDR3 variants for each isoform. Each construct has the same sequence as those in the International ImMunoGeneTics information system (IMGT) database (except the CDR3 region). The three CD3 variants from each isoform are mixed in a 16:4:1 ratio present in three discrete concentrations of 4000, 1000 and 250 RNA molecules (i.e., UMIs)/µl.
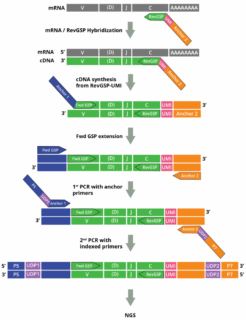
The spike-in control RNA mix has been designed to serve as universal RNA positive control for validating, calibrating, and standardizing commercial and home brew AIRR Sequencing assays using multiplex RT-PCR such as Cellecta´s DriverMap AIR Kits or 5’-RACE (SMART) assays:
• Assess Immune Receptor Profiling Assay Performance: Pre-mixed RNA Spike-In Control added to the RT-PCR Immune Profiling assay provides a calibration standard for the sequencing data to eliminate biases caused by PCR variation and the different sequencing platforms
• Ensure Sensitivity and Specificity: Spike the RNA Control Mix into your experimental sample to measure the accuracy and linear range of your PCR-based assay
• Measure Sequence Errors: Due to the enormous complexity and variability in sequences of the TCR repertoire, any biases generated in the RT-PCR, NGS, or data processing steps, can significantly affect the AIRR profiling results. Using the Spike-In RNA Control Mix with defined reference sequences is an excellent strategy to accurately measure sequence errors and evaluate mechanisms responsible for generating these errors.
• Control Sample Quality: The Spike-In RNA Control Mix may be used to assess sample quality such as in degraded FFPE samples
Product Contents:
TCR 39 RNA Premix: 100 µl (Sufficient for 50 Spike-Ins using RNA samples from whole blood, PBMC, T Cell fractions, and lymphoid tissues, and 200 Spike-Ins using RNA samples from cancer biopsies, non-lymphoid tissues, and FFPE tissue samples).
Dilution Buffer: 500 µl
PBMC Control RNA: 5 µl