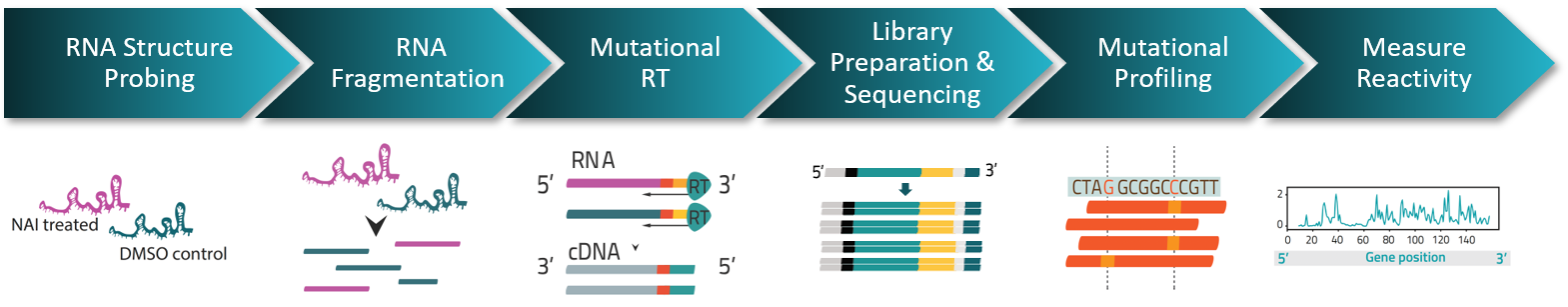
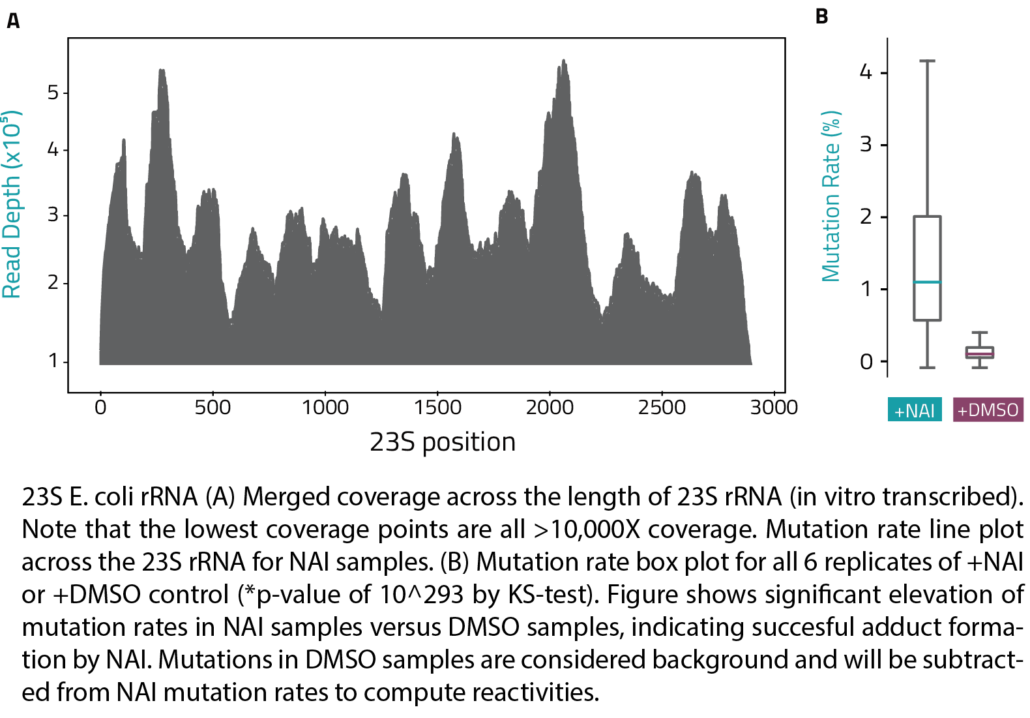
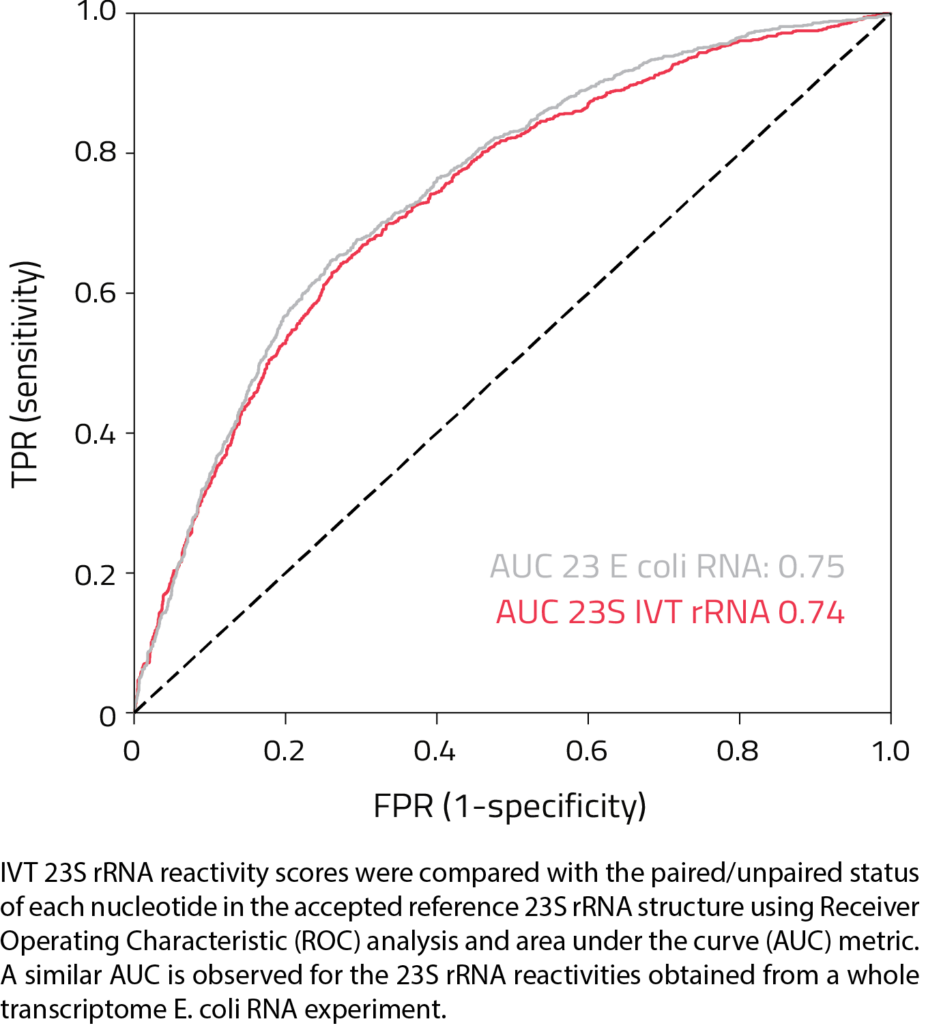
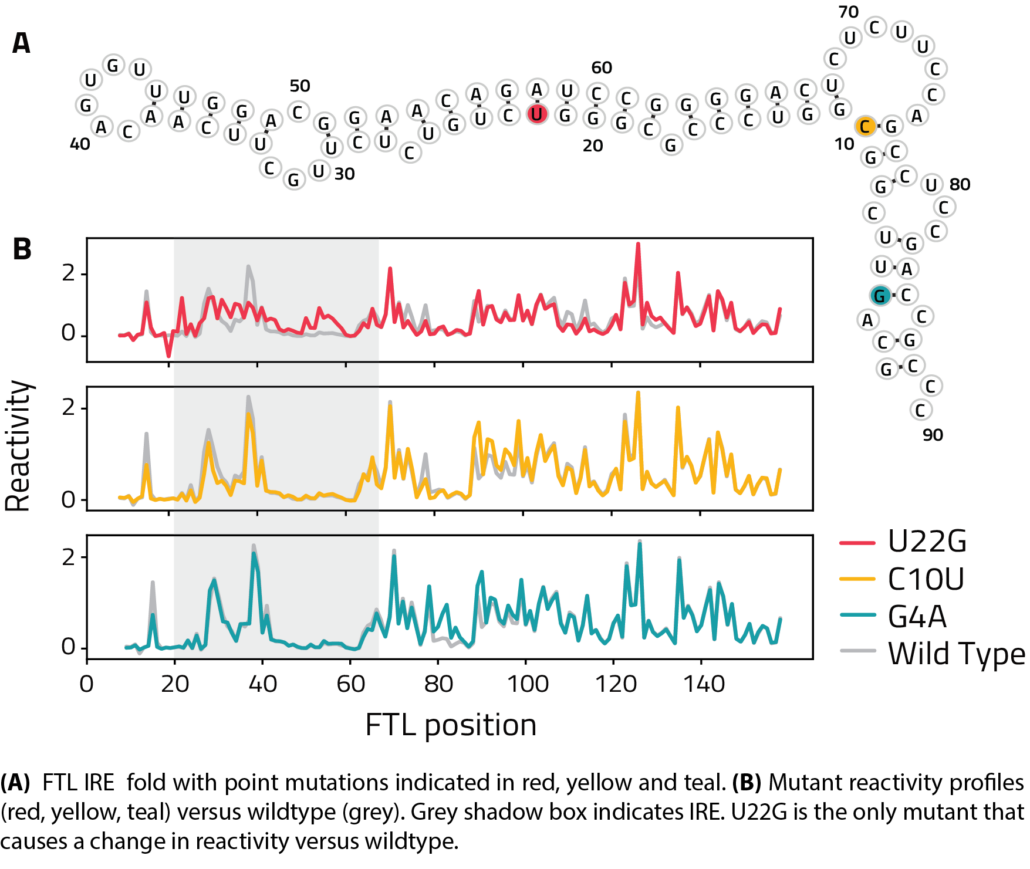
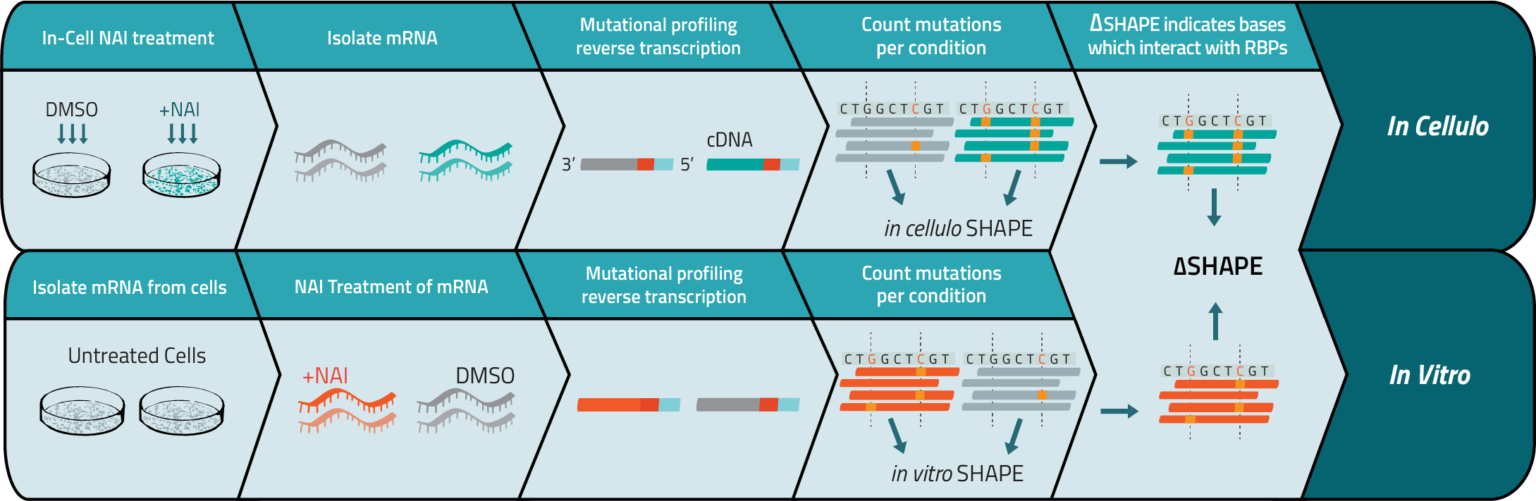
SHAPE RNA Structure Probing
- Reactivity values are predictive of paired and unpaired base status in an RNA secondary structure fold
- Compare reactivities of similar RNAs to identify regions of RNA folding variability
- Reveal differences in RNA folding across conditions or due to an outside binding factor
Showing the single result