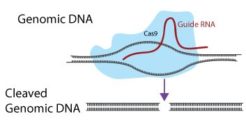
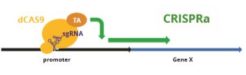
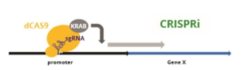
Cas9 Activity Assays
CRISPR-Test Cas9 and dCas9 Activity Test Kits
Would you like to accurately measure the activity of CRISPR Cas9 nuclease (for CRISPR KO), dCas9-KRAB (for CRISPRi), or dCas9-activator such as dCas9-VPH (for CRISPRa) in your engineered cells before starting a CRISPR screen or other CRISPR study?
Cellecta CRISPR-Test Assay Kits provide convenient FACS-based assays to accurately measure the activity of the Cas9 variant expressed by your cells. Each kit contains a mix of lentiviral particles with two different fluorescent markers. One fluorescent marker responds to the Cas9 or dCas9-variant activity measured by the kit, and the other marker provides a normalization control.
-
Measure functional activity of Cas9 nuclease, dCas9-repressor, or dCas9-activator
-
Quantitative, flow-based, read-outs
-
Convenient, complete, easy-to-use kits with ready-to-transduce lentiviral particles
Cellecta offers kits to measure different Cas9 variants using either of two different mechanisms: Fluorescent-Reporter CRISPRu/i/a-Test™ Assays directly knockout, knockdown, or induce a GFP reporter gene. Whereas, the Essential-Gene CRISPRtest™ Cas9 Assays knock out an essential endogenous gene required for viability, which leads to loss of cells with a fluorescent marker.
Fluorescent-Reporter CRISPRu/a/i-Test™ Assays: CRISPRu/CRISPRa/CRISPRi-Test Assay Kits work in any mammalian cell and typically can be run in ~4 days. They contain ready-to-transduce viral particles with a GFP construct that is responsive to the Cas9 variant being measured, and a vial of lentiviral particles constantly expressing RFP. The change of the GFP expression level relative to the RFP expression measured by FACS analysis provides an accurate measurement of the activity of the Cas9 variant expressed in the cells.
Additionally, a set of four Cas9/dCas9-fusion expression constructs is available at a reduced price when purchased in conjunction with one of the CRISPRu/a/i-Test Assay Kits.
The Essential-Gene CRISPRtest™ Cas9 Activity Assays are based on knocking out an essential gene required for cell viability. The disruption of this gene leads to loss of cells with the fluorescent marker. A construct with a non-targeting sgRNA has a second fluorescent marker, which is used for normalization. These assays are specific for a target organism and are available for human or mouse cells. Since loss of fluorescence is dependent on cell death, the assay depends on the cell growth rate and usually take 6-10 days to perform. As the assay mechanism closely mimics the knockout and cell depletion of a CRISPR screen, it is also useful to get an idea how long a screen should run. We offer these assays with various fluorescent protein readouts (GFP, RFP, BFP).
Workflow:
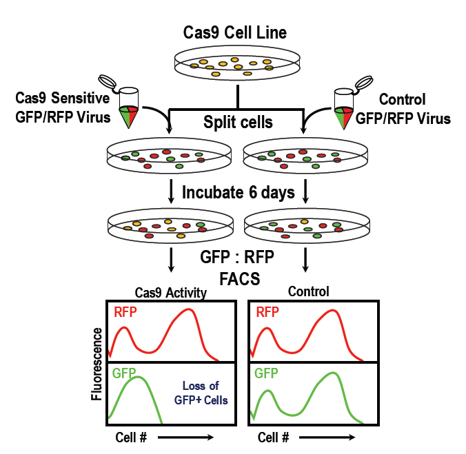
- Transduce your target population of putative Cas9, dCas9-activator, or dCas9-repressor cells with the ready-to-transduce lentiviral mix which contains fluorescently labeled lentiviral constructs, and sgRNA compatible with the Cas9 variant being measured.
- Determine the baseline fluorescence ratio of two fluorescent markers by flow cytometry.
- After several days, again measure the fluorescent cells by flow.
- Assess Cas9 or dCas9-variant activity based on the difference in the initial vs. end-point fluorescence ratios.
Showing all 11 results