Description
eSHAPE Service: SHAPE RNA Structure Probing for Single RNA provided by you
Obtain NGS-based in vitro SHAPE RNA Structure Probing data through NAI structure probing of your IVT RNA
eSHAPE single RNA is a method to obtain SHAPE RNA structure probing data for in vitro transcribed RNA formulations. In vitro transcribed or synthetic RNAs are used as input for RNA structure probing with the NAI reagent. eSHAPE single RNA is available as a kit for end user completion, and as a full service offering with results delivered as a data package containing RNA structure information as reactivity values.
• Compare similar single RNAs to identify regions of RNA folding variability
• Reveal differences in RNA folding across different conditions or due to RNA binding factors
• Extremely deep RNA coverage >10,000X with minimal sequencing depth
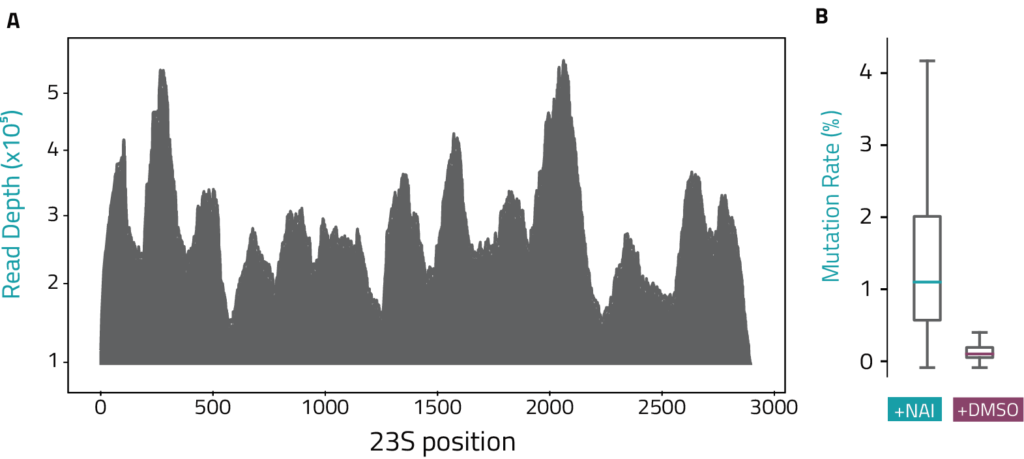
• Elevated mutation rates in NAI samples compared to the DMSO control identify RNA positions with high reactivity/unpaired bases
• Reactivity values are predictive of paired and unpaired base status in an RNA secondary structure fold
Eclipse Bio´s SHAPE RNA Structure Probing technology on IVT RNAs measures local RNA flexibility in vitro for prediction of RNA secondary structure for the RNA formulation:
Selective 2’-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) is a chemical probing method that measures RNA flexibility at single nucleotide resolution. The licensed use of the patented reagent NAI (2-methylnicotinic acid imidazolide) in the RNA probing reaction forms adducts with the free 2′-OH on the RNA backbone in single stranded regions. In contrast to other probing agents, NAI is unbiased and modifies all 4 RNA bases in single stranded RNA regions. The reverse transcription reaction of these adducts induces mutations in the newly formed cDNA at a higher rate than the DMSO control sample. Mutation rates are determined by NGS and calculated for each position along the IVT RNA. Computing into SHAPE reactivities guides the folding of the RNA by indicating the pairing status for each position.
eSHAPE Single RNA Service Description:
You provide: 1 ug in vitro transcribed or synthetic RNA
The service comprises:
• Probing of Your RNA with NAI and control incubation of your RNA with DMSO
• Construction of two eSHAPE NGS Libraries: 1 NAI NGS Library and 1 DMSO NGS Library
• Sequencing of both libraries on Illumina: 8M Reads (SE100) each and subsequent comprehensive data analysis
Deliverable:
You receive the following comprehensive and complete eSHAPE Single RNA Data Analysis Package:
⋅ RAW SEQUENCING DATA | FASTQ:
Reads directly off sequencer and demultiplexed per sample for data back-up and publication
⋅ RNA ALIGNMENTS | BAM:
Reads aligned to the RNA of interest, and PCR deduplicated for additional downstream analysis
⋅ MUTATION RATES | TSV:
Mutation rates for each NAI and DMSO sample per RNA position for reactivity calculations
⋅ eSHAPE REACTIVITY | BEDGRAPH, SHAPE:
IQR normalized reactivity score at each position of the RNA of interest with >1000x coverage for visualization in a genome viewer such as IGV or for use as input into the RNAStructure algorithm to guide fold predictions
⋅ eSHAPE SUMMARY REPORT | HTML:
Report of eSHAPE results with interactive tables and plots for publication ready graphics. Includes QC metrics, coverage, mutation rates and reactivities for the RNA of interest
⋅ REACTIVITY COMPARISON REPORT | HTML:
Report of differential reactivities for experiments with multiple conditions or RNAs for comparison. Includes interactive plots and results table denoting significantly different positions with p-values
We are offering scaled pricing depending on the number of RNA samples provided –
please contact us for a quote!