Description
3´ UTR Sequencing Service for detecting 3´UTR Variants by constructing mRNA 3´-end enriched RNA-Seq NGS libraries
Reveal 3′ UTR Landscape and Poly Adenylation Signals and Sites genome-wide by 3´ UTR Sequencing
3′ End-Sequencing is a technology for defining 3′ ends of transcripts by next generation sequencing. 3′ End-Sequencing can identify with high confidence known and novel Poly Adenylation signals and sites at single nucleotide resolution.
• Define 3′ UTR Landscape: Genome-wide detection of known and novel poly adenylation sites (PAS) to define 3′ UTRs.
• Single Nucleotide Resolution: PolyA+ sites are detected with single nucleotide resolution genome-wide.
• High Confidence Poly Adenylation Sites: No calling of sites that map to genomic A-rich regions
• High Throughput and Robust Workflow
• High Reproducibility with accurate data
• Unbiased with high specificity
Poly Adenylation Sites (PAS) and the 3′ UTR are involved in mRNA regulation and stability, and are implicated in disease and can function as disease biomarkers and drug targets.
3´End-Seq facilitates precise UTR identification for RNA therapeutics targeting and can be used as a tool for identifying UTR biomarkers associated with disease. 3′ UTR Sequencing can quantify the relative usage of PAS across samples and indicate transcript isoform presence in annotated transcriptomes. Defining the 3′ UTR with 3´End-Seq also helps predict binding factor motifs more reliably.
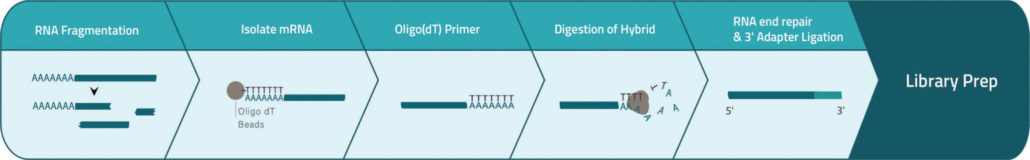
3′ End-Seq Workflow
Total RNA is heat fragmented to 100-200 nucleotide fragments and polyA (+) mRNA fragments are enriched by double oligo(dT) bead capture. Subsequently oligo(dT) primers are added to poly(A)+ mRNA. The hybridized polyA tails are trimmed away enzymatically and after a clean-up step an RNA-seq library covering the highly enriched antisense-strand mRNA 3´ end fragments is prepared.
Sample Input: >3 ug Total RNA (>0.1 ug/ul), RIN >7
Sequencing Recommendations: 10-15M reads (SE100) on Illumina
3′ End-Seq Service Description
You provide:
10 µg Total RNA; RIN ≥ 3; Non-Crosslinked
The service comprises:
- mRNA Enrichment
- PolyA Tail Digestion
- Adapter Ligation
- Library preparation
- Sequencing: 10-15M reads and comprehensive data analysis
Deliverable:
You receive the following comprehensive Data Analysis Package:
⋅ RAW SEQUENCING DATA | FASTQ:
Reads directly off sequencer and demultiplexed per sample for data back-up and publication
⋅GENOME ALIGNMENTS| BAM:
Reads filtered of repetitive elements, aligned, and PCR deduplicated for additional downstream analysis
⋅COVERAGE TRACKS| BIGWIG:
Normalized read end coverage on positive and negative strands for visualization in a genome viewer such as IGV
⋅SINGLE NUCLEOTIDE 3´ END CALLS| BED:
Genomic single nucleotide positions scored as putative 3’ transcript ends for 3’UTR exploration
-3′ End-Seq SUMMARY REPORT | HTML
Report with interactive tables and plots for publication ready graphics. Includes QC metrics and 3’ end enrichment
We are offering scaled pricing depending on the number of RNA samples provided –
please contact us for a quote!