Categories
CloneTracker XP 5M Barcode-3′ Library (packaged)-5 pools (pScribe4M-RFP-Puro)
22.100,00 €
Quantity:
5 x 10^8 TU
Cat.#: BCXP5M3RP-5S-V
Supplier: Cellecta
Categories: Cell Barcoding, RNA-Seq Barcode Pooled Libraries
Tags: Cell Lineage Tracing, scRNA-seq Barcodes, RNA-seq Barcodes, NGS Barcodes, Clonetracker XP, Drop-Seq Barcode Library, scRNA-Seq Barcode Library, RNA-Seq Barcode Library, Single Cell Analysis, Lineage Tracing, Clonetracker, Cell Labeling, Clonal Tracking, Cell Tracking, Lentiviral Barcode Library, DNA Barcode Library, NGS Barcode Library, Cell Barcoding, Cellular Barcoding
Description
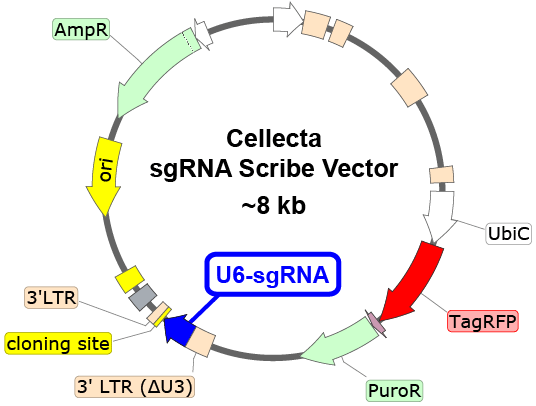
Each 5M barcode validated, pooled lentiviral library can be used to easily label ~ 1 million cells with heritable unique RNA-expressed barcodes, detectable by RNA-Seq and single-cell analysis as well as in the genomic DNA fraction. The barcodes are non-overlapping enabling to label each cell population individually. These library pools all incorporate the barcode in the 3′-UTR of a transcript, which is compatible with oligo-dT cDNA synthesis protocols.
Shipping Conditions:
Dry Ice
Storage Temperature:
-80C