Description
Profile T-Cell Receptor (TCR) adaptive immune receptor repertoire (full-length variable region) by AIRR-seq starting from human total RNA
Easy-to-run, single-day AIRR-seq assay that uses multiplex PCR-NGS technology to profile adaptive immune receptor repertoires
• Only assay technology on the market that profiles the human CDR1 CDR2 CDR3 repertoire of all T-Cell Receptor (TCR) and B-Cell Receptor (BCR) chains in a single-tube AIRR-seq assay
• Start from various immune sample types such as whole blood, PBMC, cancer biopsies, tissue samples, FFPE or dried blood microsamples.
• Detect and accurately quantify more AIR clonotypes by Unique Molecular Identifiers (UMIs)
• Amplify only functional TCR/BCR mRNA isoforms, without pseudogenes, ORFs or misarranged genes
• Run in parallel with the DriverMap AIR TCR and BCR kits with DNA as starting material for insights into activated clonotypes
Cellecta’s DriverMap™ AIR Adaptive Immune Receptor Repertoire Profiling assay is a next-generation targeted multiplex RT-PCR NGS technology designed for bulk expression profiling in a single assay of all functional T-Cell Receptors (TCR) and B-Cell Receptors (BCR): TRA, TRB, TRD, TRG, IGH, IGK and IGL mRNA isoforms excluding non-functional pseudogenes and ORFs.
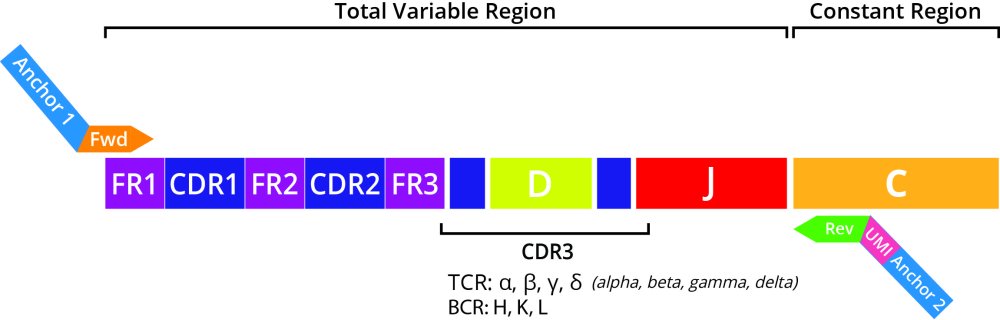
DriverMap AIR full-length variable region assays allow profiling the repertoire of full-length receptor regions (CDR1, CDR2, and CDR3) using a set of experimentally validated forward primers (designed for variable FR1 region), and reverse primers (designed for the conserved C region) of TCR and BCR mRNAs. Two sets of AIR primers specific for TCR (TRA, TRB, TRD, and TRG) and BCR (IGH, IGL, and IGK) chains are offered separately in AIR reagent kits to allow profiling AIR repertoires individually for TCRs or BCRs or together in a single reaction. Profiling the full-length receptor region gives a complete picture of all the CDR1, CDR2, and CDR3 variable region repertoire diversity which is useful for understanding epitope recognition for T-cells and antibody discovery applications for B cells.
While the kit described here profiles the T-cell chains only, we recommend profiling both TCR and BCR repertoires together, since in most cases, T- and B-cells work synergistically in the adaptive immune response.
Kit Description and Contents:
• Profile all TCR chains (TRA, TRB, TRD, and TRG) in a single reaction from a single sample.
• Includes all the enzymes, reagents, and primers to prepare 24 samples ready for NGS from human total RNA.
• Assay(s) can be run in a single reaction without having to run multiple expensive assays and without any specialized equipment
• Generate interactive reports for clonotype quantitation, CDR1-3 length distribution, V(D)J segment usage, and isotype composition using MiXCR (MiLabs) adaptive immune receptor repertoire profiling software.