Description
High Quality NGS Library Prep including methylated CpG enrichment using 20 ng – 500 ng Bisulfite Treated DNA inputs
• Enrichment of methylated CpG sites
• Single-base resolution
• Low cost for sequencing
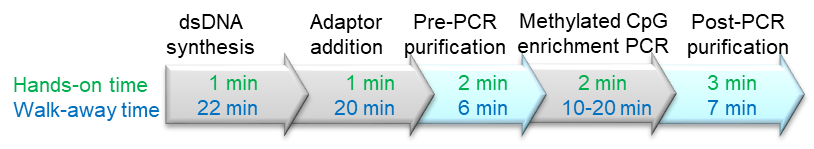
• Fast: Total time: 1.5 hours, Hands-on time: 10 minutes
• Easy procedure
• Less magnetic beads required: Reduced more than 50%
• Guaranteed high bisulfite sequencing library conversion efficiency
• Input DNA: 20 ng to 500 ng bisulfite treated DNA
The Methylation Specific Bisulfite-Seq (MSBS) Library Prep Kit (illumina platform) was developed for construction of NGS libraries for methylated CpG sites using bisulfite treated DNA (20 ng – 500 ng) as input. DNA methylation is important for regulation of cell development, differentiation and gene expression in molecular biology, genetics and epigenetics. Most methylated cytosines are found at CpG sites, and 70-80% of cytosines are methylated. The number of CpG sites in human genome is around 28 million, which is less than 1% of the genome. This kit enriches methylated CpG regions, thus significantly reducing sequencing costs compared to Whole Genome Bisulfit Sequencing. The kit detects the whole genome methylation patterns at the single base level since it is based on bisulfite-seq technology. DNA methylation is an epigenetic mechanism known to play a critical role in gene regulation and genomic imprinting by blocking transcription factor access to promoters and enhancers. Bisulfite sequencing is a popular technique in biomedical research used to detect and analyze methylation markers at a single base pair resolution. Treating DNA with sodium bisulfite converts unmethylated cytosine into uracil via deamination, while methylated cytosine remains in its original form.
Recently, NGS became a powerful tool to identify the DNA methylation status at the whole genome level with single-base resolution. However, it is well known that bisulfite treatment of the NGS libraries causes tremendous damage to the libraries. In the case of the regular bisulfite seq library preparation (library prep before bisulfite conversion), DNA shearing equipment and the expensive methylated adaptors are required. In addition, the subsequent bisulfite conversion causes tremendous DNA damage to the constructed libraries.
BioDynami has developed a unique library prep technology to solve the problems. The technology uses bisulfite treated DNA as input to avoid the significant library loss caused by bisulfite conversion. Furthermore, DNA shearing step and expensive methylated adaptors are not required with the kit. The DNA polymerase in the kit has high-fidelity amplification ability and uracil tolerance which is ideal for amplification of bisulfite sequencing libraries.
The workflow of the bisulfite DNA kit is simple: make the libraries in two steps followed by PCR and cleanup steps. The fast and simple 1.5-hour protocol makes libraries with even coverage and low GC-bias based on BioDynami´s unique chemistry for DNA end-polishing and adaptor addition.
Library multiplexing is possible with different types of indexes.
With BioDynami’s unique DNA library preparation technologies, the fast and simple NGS DNA Library Prep Kit allows high quality NGS library preparation to be completed in 1.5 hours with only 10 minutes of hands-on time.
Standard beads purification can be used to clean the constructed libraries after the library prep and subsequent PCR amplification step. The optimized beads purification step not only reduces the working time, but also decreases more than half of the beads amount needed.
Different Kit versions are available as follows
Non-index (Cat.# 30101): Libraries do not have index.
Index (Cat.# 30102): Each of our index primers contains a unique barcode sequence with 6 bases that can be used to identify libraries. Library multiplexing up to 48 samples is possible.
Unique dual index (Cat.# 30103): Library multiplexing up to 96 samples is possible with unique dual indexes. We have developed a 4-Base Difference Index System. The system allows us to make indexes that have at least 4 bases different from each other in the 8 bases index length. Our unique dual indexing primers remove sequencing errors such as index hopping, index cross-contamination, mis-assignment of reads, amplification errors, and de-multiplexing errors. The primer set includes 96 pre-mixed unique pairs of i5 and i7 index primers in a 96-well plate