Description
Profile T-Cell Receptor (TCR) adaptive immune receptor repertoire by AIRR-seq starting from human genomic DNA
Easy-to-run, single-day AIRR-seq assay that uses multiplex PCR-NGS technology to profile adaptive immune receptor repertoires
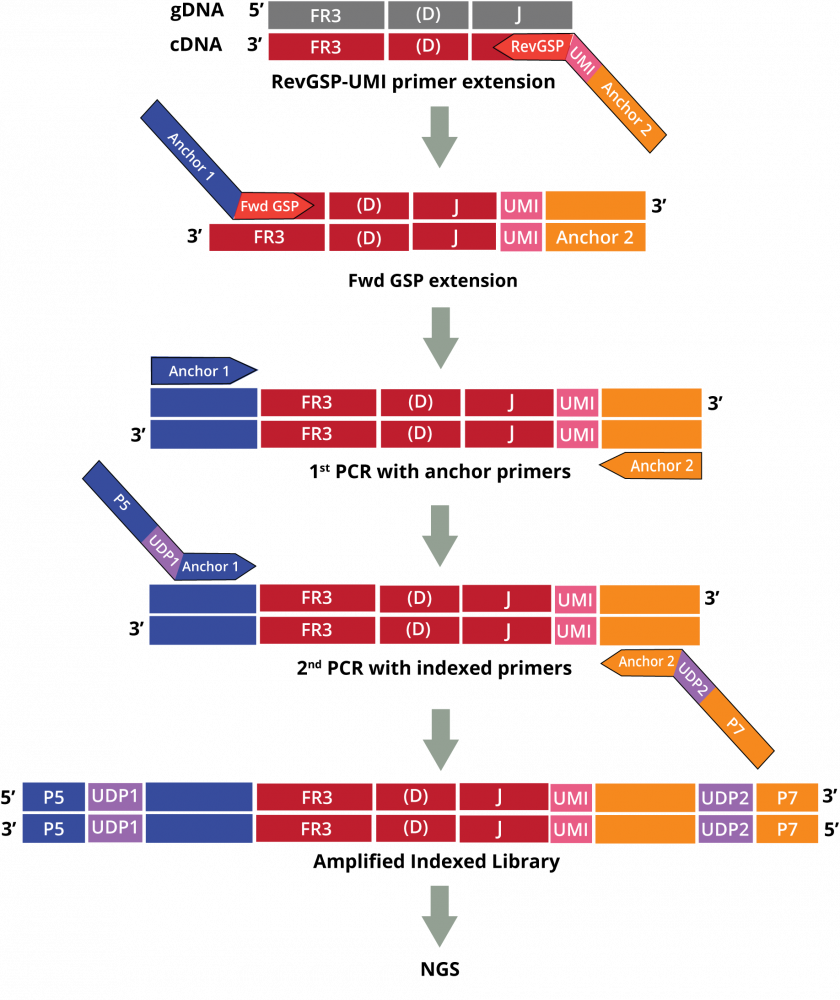
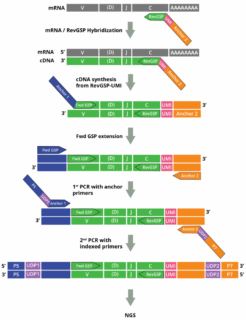
The DriverMap AIR DNA assay measures the frequency of CDR3 clonotypes to quantify the clonal frequency of T cells.
• Assay primers contain unique molecular identifiers (UMIs) to enable accurate quantitation of each clonotype.
• The concentration of reverse gene-specific primers is optimized to provide balanced amplification efficiency of all chains for TCR (TRA, TRB, TRD, and TRG)
• Sensitive profiling of samples with low lymphocyte content (e.g. FFPE, tumor biopsy, archived specimens) where DNA is more stable than RNA.
• Complete kit includes all the enzymes, reagents, and primers to prepare 24 samples ready for multiplex Illumina NGS runs from DNA starting material. No additional specialized equipment required.
• Parallel runs of DriverMap AIR DNA Assay with DriverMap AIR RNA Assay reveal potential disease-associated, activated CDR3 clonotypes
Cellecta’s DriverMap™ AIR Adaptive Immune Receptor Repertoire Profiling assay is a next-generation targeted multiplex RT-PCR NGS technology designed for bulk quantification of all functional T-Cell Receptors (TCR) TRA, TRB, TRD, TRG in a single AIRR-seq assay.
Kit Description and Contents:
• Profile all TCR chains (TRA, TRB, TRG and TRD) in a single reaction from a single sample.
• Includes all the enzymes, reagents, and primers to prepare 24 samples ready for NGS from human genomic DNA.
• Assay(s) can be run in a single reaction without having to run multiple expensive assays and without any specialized equipment
• NGS data can be analyzed with the MiXCR software package available online