Description
Profile T-Cell Receptor (TCR) adaptive immune receptor repertoire starting by AIRR-seq from human total RNA
Easy-to-run, single-day AIRR-seq assay that uses multiplex PCR-NGS technology to profile adaptive immune receptor repertoires
• Only assay technology on the market that profiles the human CDR3 repertoire of all T-Cell Receptor (TCR) and B-Cell Receptor (BCR) chains in a single-tube AIRR-seq assay
• Start from various immune sample types such as whole blood, PBMC, cancer biopsies, tissue samples, FFPE or dried blood microsamples.
• Detect and accurately quantify more AIR clonotypes by Unique Molecular Identifiers (UMIs)
• Amplify only functional TCR/BCR mRNA isoforms, without pseudogenes, ORFs or misarranged genes
• Run in parallel with the DriverMap AIR TCR and BCR kits with DNA as starting material for insights into activated clonotypes
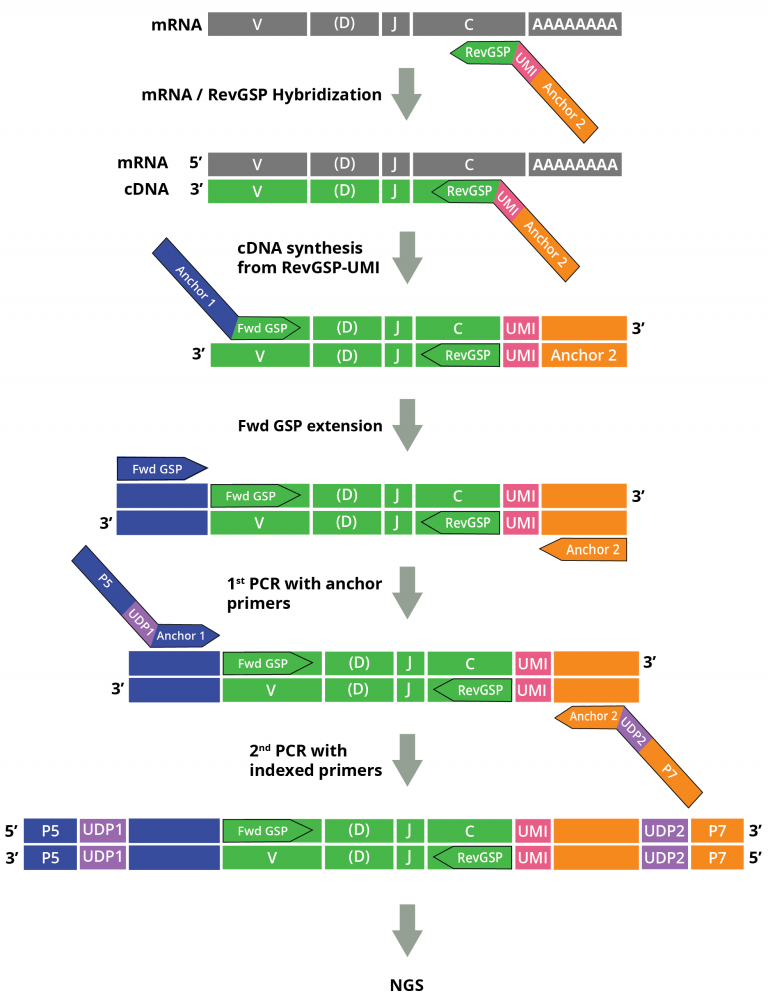
Cellecta’s DriverMap™ AIR Adaptive Immune Receptor Repertoire Profiling assay is a next-generation targeted multiplex RT-PCR NGS technology designed for bulk expression profiling in a single assay of all functional T-Cell Receptors (TCR) and B-Cell Receptors (BCR): TRA, TRB, TRD, TRG, IGH, IGK and IGL mRNA isoforms excluding non-functional pseudogenes and ORFs.
While the kit described here profiles the T- cell chains only, we recommend profiling both TCR and BCR repertoires together, since in most cases, T- and B-cells work synergistically in the adaptive immune response.
Kit Description and Contents:
• Profile all TCR chains (TRA, TRB, TRG and TRD) in a single reaction from a single sample.
• Includes all the enzymes, reagents, and primers to prepare 96 samples ready for NGS from human total RNA.
• Assay(s) can be run in a single reaction without having to run multiple expensive assays and without any specialized equipment
• Generate interactive reports for clonotype quantitation, CDR3 length distribution, V(D)J segment usage, and isotype composition using MiXCR (MiLabs) adaptive immune receptor repertoire profiling software.