ProxiMeta Platform: ProxiMeta Hi-C Kit for Microbial Samples and ProxiMeta Data Analysis Package
4.750,00 €
Description
Proximity Ligation NGS Library Prep from crude microbial samples, for Illumina Sequencing, plus online metagenome deconvolution analysis of your sequence data
.
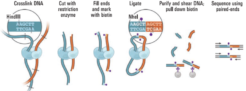
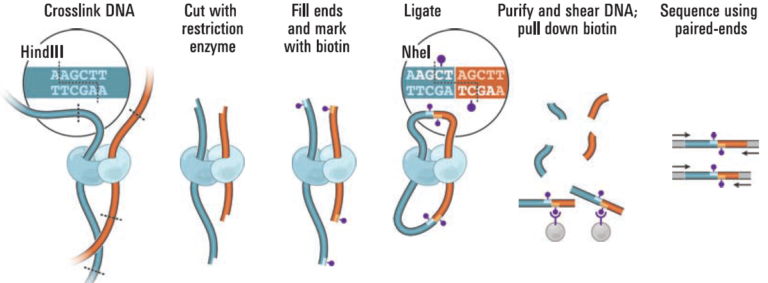
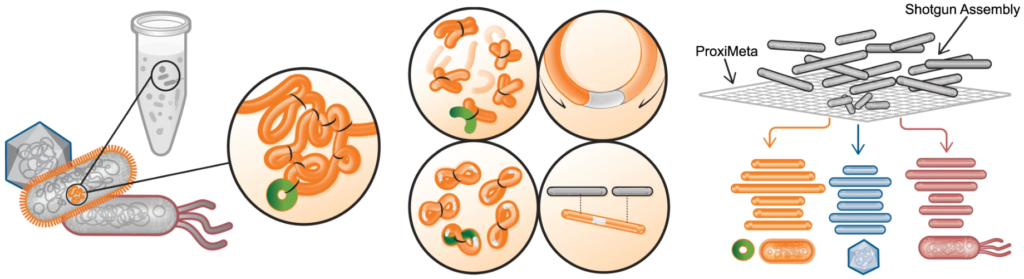
Proximity ligation (Hi-C) libraries are generated from a single mixed microbial sample. Interactions are captured by crosslinking, digesting, and creating chimeric junctions that are sequenced.
The Hi-C sequence data is analyzed on a cloud-based bioinformatics platform along with a shotgun assembly to deconvolve chromosomes and mobile genetic elements into complete genomes.
• Assemble hundreds of high-quality genomes directly from the microbiome
• No high-molecular-weight DNA or culturing required
• Associate mobile genetic elements (MGEs) such as transposons, phages, plasmids, and antimicrobial resistance genes (ARGs) with their hosts
• Discover novel organisms, strains, and genes
• Short-read compatible; yields libraries for Illumina® sequencing
• User-friendly complete 8-pack Hi-C Kit including NGS library prep reagents and eight unique dual index primers for multiplexing, plus cloud-based ProxiMeta deconvolution analysis
Phase Genomics´ ProxiMeta Metagenome Deconvolution Platform represents a paradigm shift in metagenome and microbiology research, enabling to obtain lineage-resolved, complete metagenome-assembled genomes from complex microbial communities. It combines kit-generated proximity ligation data with shotgun sequencing data, to assemble high-quality metagenomes and associate mobile genetic elements with their hosts. Now you can capture deep strain-resolution insights and analyze bacteriophage, plasmid, and transposon attributions to their hosts (see application note).
Assemble hundreds of high-quality genomes without guesswork
Unlike 16S rRNA analysis or binning approaches, proximity ligation provides direct, physical evidence of sequences co-located in the same cell. ProxiMeta uses this information to cluster proximity-assembled genomes (PAGs) for both eukaryotic and prokaryotic members of the microbiome population. Each genome is assessed for completeness, contamination and novelty using industry-leading computational tools.
Link transposons, plasmids and antibiotic resistance genes to their hosts
In addition to genome assemblies for the members of a metagenomic sample, the ProxiMeta workflow also yields sequence information for mobile genetic elements (MGEs). These may include plasmids, transposons and antibiotic resistance genes (ARGs). Since proximity ligation is performed on intact cells, MGE-host interactions are also captured. During the ProxiMeta assembly, mobile genetic elements are associated with their hosts allowing you to gain insight into horizontal gene transfer in complex microbial communities.
Assign phages to their hosts
During the proximity ligation library prep, phage and viral DNA is crosslinked to the DNA of bacterial hosts within a mixed microbial sample. Those interactions are captured in the sequences of chimeric junctions, which allows the ProxiMeta algorithms to accurately assign phages to their hosts during the deconvolution analysis. Phage-host attribution produces a more accurate view of the microbiome from a single Hi-C library.
Discover more … species, strains, genes and gene functions
Moreover, the ProxiMeta™ Metagenome Deconvolution Platform is designed for discovery and bioprospecting, as it does not require culturing or rely on any a priori data. With the ability to assemble high-quality genomes for microbes with as little as 0.05% cellular abundance in a sample, it is easy to discover novel organisms, strains and genes, and predict gene function.
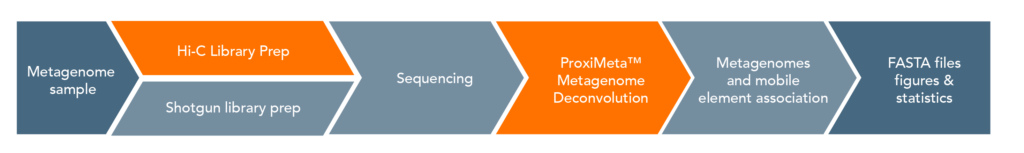
ProxiMeta Workflow:
The ProxiMeta platform provides a sample-to-analysis solution that starts with a crude metagenomic sample, and ends with analyzed data. The entire process is typically completed in a month.
The proximity ligation (Hi-C) library prep reagents provide a streamlined protocol that does not require culturing of microbes or extraction of high-molecular weight (HMW) DNA. Processing guidelines for different types of metagenomic samples ensure robust results. The 8-step library prep workflow can be completed in 2 working days, and requires only 3 hours of hands-on time. The protocol contains several convenient, safe stopping points.Sequencing may be performed on any Illumina® sequencer. The ProxiMeta Kit yields a dual-indexed proximity ligation library, for which ~50 to 100 million paired-end reads are required. For analyzing your Hi-C sequence data, you also need to provide shotgun sequencing data of your samples, which may be performed with a suitable library preparation kit of your choice – contact us for recommendations or to discuss a service project if you do not have prior experience with shotgun library preparation and sequencing.
ProxiMeta Metagenome Deconvolution: Full Data Analysis Package included in the Kit:
Metagenome deconvolution analysis of your sequence data including linkage to plasmids, antibiotics resistance genes, and assignment of phages to their hosts is included in this combination package product. You can perform comprehensive data analysis using the interactive cloud based ProxiMeta™ Explorer.
Click here to experience the ProxiMeta Explorer Data Analysis Platform with several published genome-resolved metagenomic datasets.